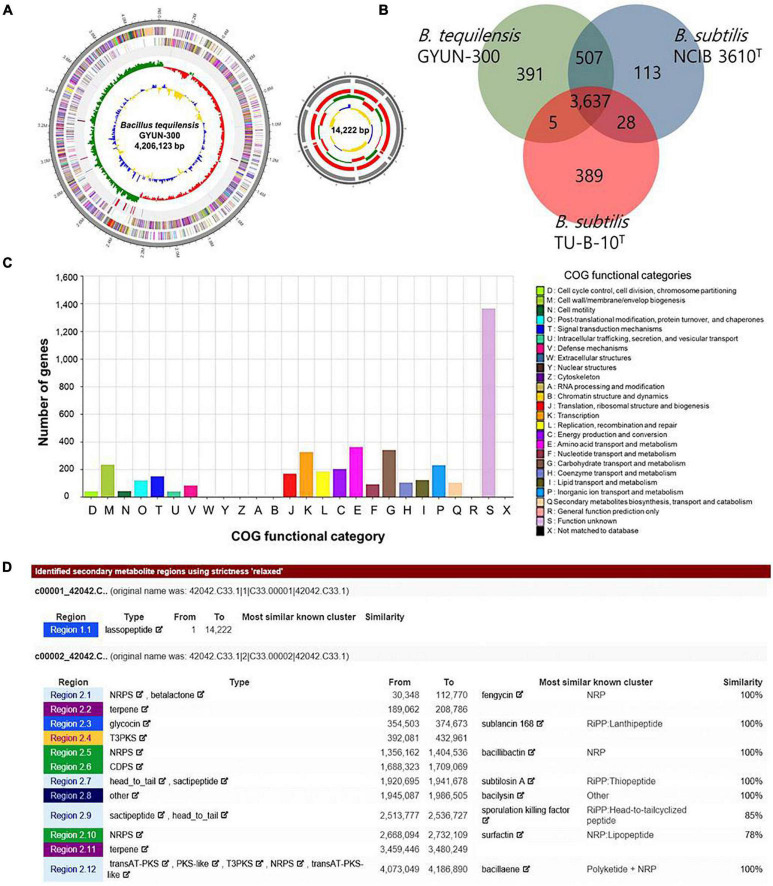
FIGURE 5.
Whole-genome map of Bacillus tequilensis GYUN-300 and annotation. (A) Marked characteristics shown from the outside to the center; coding sequence (CDS) on forward strand, CDS on reverse strand, tRNA, rRNA, guanine-cytosine (GC)-content, and GC skew. CDS genome consists of a single circular chromosome that is 4.2 mb in size, and represents one contig from the outer part of the circle; the second circle represents forward, the third represents reverse strain, and the fourth circle represents tRNA and rRNA positions; (B) distribution of orthologous genes in the Bacillus tequilensis GYUN-300, B. subtilis subsp. subtilis NCIB 3610T, and B. subtilis subsp. spizizenii TU-B-10T genomes. The Venn diagram shows the summary of unique SNPs from total genes of the B. tequilensis GYUN-300 strain. This analysis exploits all CDS of the genomes and is not restricted to the core genome; (C) the clusters of orthologous genes (COG) function annotation of B. tequilensis GYUN-300, and distribution of genes in different COG function categories; and (D) analysis of secondary metabolites of GYUN-300 using the antiSAMASH program.