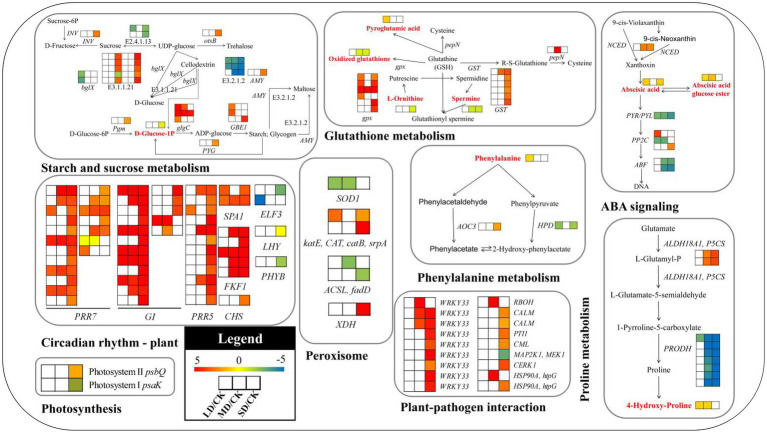
Figure 9.
Key metabolic pathways and candidate genes in S. davidii associated with the three treatment groups under drought stress. E3.2.1.2, beta-amylase; E2.4.1.13, sucrose synthase; E3.1.1.11, pectinesterase; E3.2.1.21, beta-glucosidase; glgC, glucose-1-phosphate adenylyltransferase; GBE1, 1,4-alpha-glucan branching enzyme; bglX, beta-glucosidase; Pgm, phosphoglucomutase; PYG, glycogen phosphorylase; INV, beta-fructofuranosidase; AMY, alpha-amylase; otsB, trehalose 6-phosphate phosphatase; gpx, glutathione peroxidase; pepN, aminopeptidase N; GST, glutathione S-transferase; PP2C, protein phosphatase 2C; PYL, abscisic acid receptor PYR/PYL family; ABF, ABA-responsive element binding factor; NCED, 9-cis-epoxycarotenoid dioxygenase; PRR7, pseudoresponse regulator 7; PRR5, pseudoresponse regulator 5; SPA1, protein suppressor of PHYA-105 1; ELF3, protein EARLY FLOWERING 3; CHS, chalcone synthase; FKF1, flavin-binding kelch repeat F-box protein 1; GI, GIGANTEA; LHY, MYB-related transcription factor LHY; PHYB, phytochrome B; SOD1, superoxide dismutase; katE, CAT, catB, srpA, catalase; ACSL, fadD, long-chain acyl-CoA synthetase; XDH, xanthine dehydrogenase/oxidase; HPD, 4-hydroxyphenylpyruvate dioxygenase; AOC3, primary-amine oxidase; PRODH, proline dehydrogenase; ALDH18A1, P5CS, delta-1-pyrroline-5-carboxylate synthetase; HSP90A, htpG, molecular chaperone HtpG; WRKY33, WRKY transcription factor 33; RBOH, respiratory burst oxidase; CALM, calmodulin; PTI1, pto-interacting protein 1; CML, calcium-binding protein; MAP2K1, MEK1, mitogen-activated protein kinase kinase 1; CERK1, chitin elicitor receptor kinase 1; psbQ, photosystem II oxygen-evolving enhancer protein 3; and psaK, photosystem I subunit X. The log2[fold-change (FC)] colour scale ranges from −5 to 5, with blue indicating downregulation and red indicating upregulation (see the colour set scale in the bottom left corner). The red characters represent the metabolites in the pathway diagram.