Figure 1.
Nfatc1 expression labels a small population of mammary stem/progenitor cells (see also Figure S1)
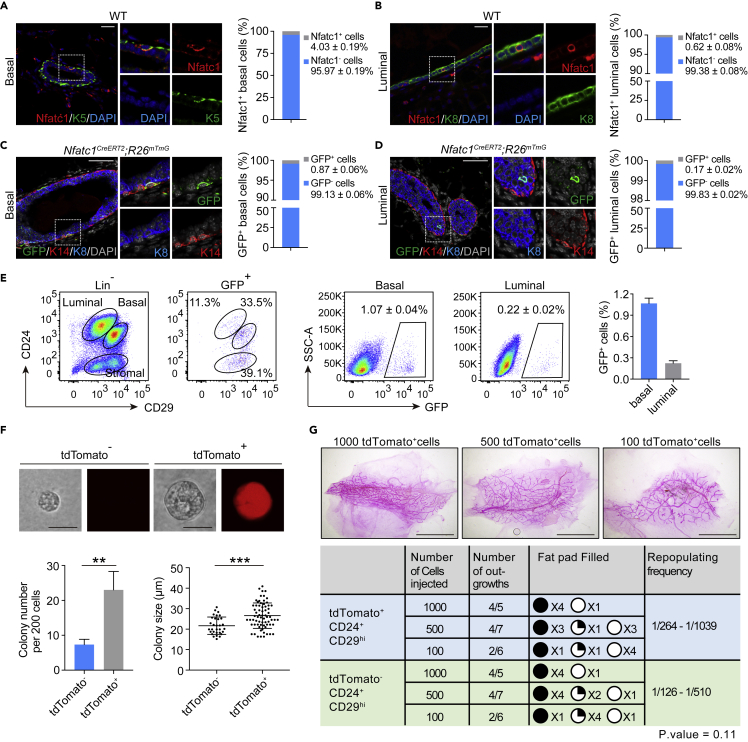
(A and B) Co-immunofluorescence assay for Nfatc1 and K5 (A) or Nfatc1 and K8 (B) in eight-week-old WT mammary glands. n = 3 mice. Scale bar, 25 μm. High-magnification images are shown on the right. The percentages of Nfatc1+ basal cells among total basal cells (A) and Nfatc1+ luminal cells among total luminal cells (B) were quantified. A total of 1,952 basal cells and 8,335 luminal cells from three mice were counted
(C and D) Co-immunofluorescence assay for GFP (green), K14 (red), and K8 (blue) in mammary glands from Nfatc1CreERT2;R26mTmG mice at the age of eight weeks. GFP+ cells were present in the basal (C) and luminal (D) layers. n = 3 mice. Scale bar, 50 μm. Quantification analysis shows the percentages of GFP+ basal cells among total basal cells (C) and GFP+ luminal cells among total luminal cells (D). A total of 1,614 basal cells and 5,314 luminal cells from three mice were quantified
(E) Flow cytometry assay of GFP+ cells from Nfatc1CreERT2;R26mTmG mice at the age of eight weeks. The mice were administered TAM three times, and the samples were collected 48 h after the last induction. Quantification analysis showing the percentages of GFP+ basal cells among total basal cells and GFP+ luminal cells among total luminal cells. Another analysis approach is also shown. Total GFP+ cells were applied to CD24 and CD29 gates, showing the distribution of GFP+ cells in the basal and luminal layers. n = 3 mice
(F) Colony formation efficiency and colony size in the Matrigel culture. For tdTomato+ and tdTomato− basal cells, a total of 69 and 31 clones were analyzed from three independent experiments. Scale bar, 25 μm
(G) Transplantation of tdTomato+ and tdTomato− basal epithelial cells by limiting dilution. Data are pooled from two independent experiments. Scale bar, 5 mm. The repopulation frequency was calculated using the method in http://bioinf.wehi.edu.au/software/elda/. Data represent the mean value ±SD ∗∗p < 0.01, ∗∗∗p < 0.001