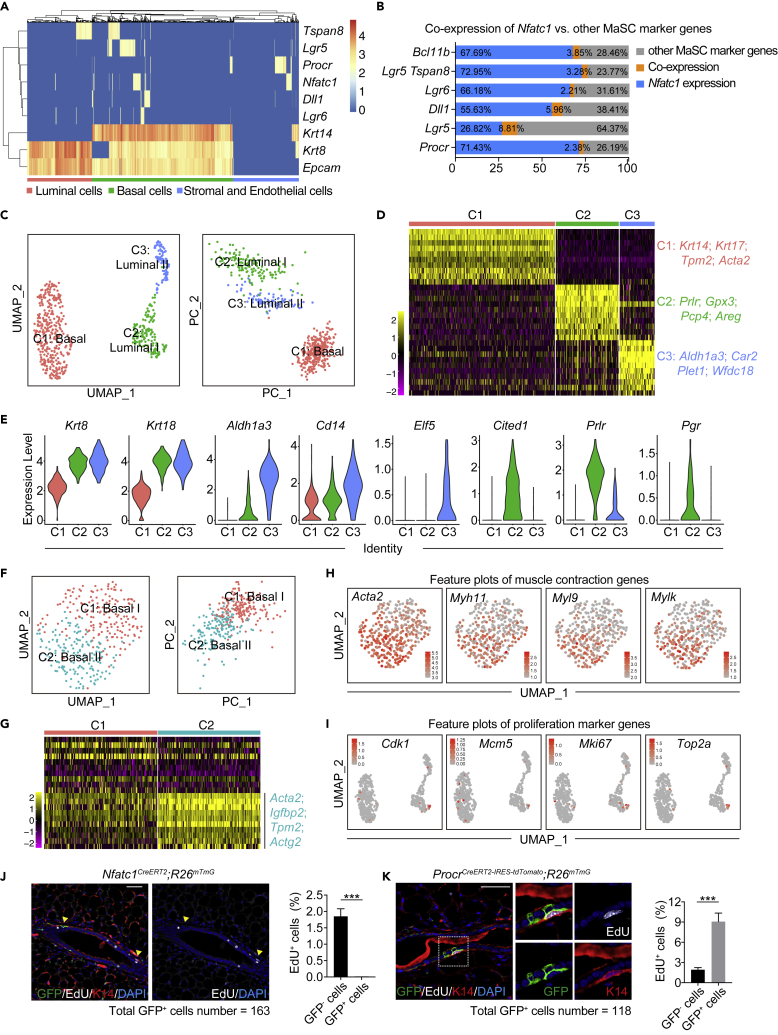
Figure 2.
Nfatc1 repotrer-marked stem/progenitor cells are dormant and distinct from other known MaSCs (see also Figure S2)
(A) Heatmap analysis of the scRNA-seq database shows the expression patterns of Nfatc1 and other MaSC marker genes (Tspan8, Lgr5, Procr, Dll1, and Lgr6), as well as Krt14 (K14), Krt8 (K8), and Epcam. The scRNA-seq database for mammary epithelial cells was adapted from a previously published study in Nature and downloaded from https://tabula-muris.ds.czbiohub.org. A nUMI count >150 was used as a cutoff for low-expression cells
(B) Quantification for the percentages of basal cells expressing Nfatc1 or the MaSC marker genes Bcl11b, Tspan8/Lgr5, Lgr6, Dll1, Lgr5, and Procr in (A). Blue indicates Nfatc1+cells; grey indicates other MaSC marker genes; and orange indicates double-positive cells
(C) UMAP and PCA plots reveal cellular heterogeneity of 558 tdTomato+ mammary epithelial cells that were sorted from Nfatc1CreERT2;R26tdTomato mice
(D) Heatmap showing differentially expressed genes in each cluster for (C). Selected signature genes are shown on the right
(E) Violin plots show marker genes of common luminal cells (Krt8/K8 and Krt18/K18), milk lineage cells (Cd14, Aldh1a3, and Elf5), and ER+ lineage cells (Cited1, Prlr, and Pgr) for each cluster in (C)
(F) UMAP and PCA plots showing the reclustering results for 333 tdTomato+ basal cells. Luminal epithelial cells were removed
(G) Heatmap of differentially expressed genes in each cluster for (F). Selected signature genes are shown on the right
(H) Feature plots showing expression distribution of genes functioning in muscle contraction
(I) Feature plots showing expression distribution of genes functioning in cell proliferation in each cluster for (C)
(J and K) Co-immunofluorescence for GFP, EdU, and K14 in Nfatc1CreERT2;R26mTmG mice (J) and ProcrCreERT2-IRES-tdTomato;R26mTmG mice (K) at 48 h post-TAM induction. Scale bar, 50 μm. The samples were collected 3 h after one dose of EdU injection. Quantification analysis shows the percentages of GFP+EdU+ cells versus GFP+ cells and GFP−EdU+ cells versus GFP− cells in Nfatc1CreERT2;R26mTmG mice (J) and ProcrCreERT2-IRES-tdTomato;R26mTmG mice (K). For Nfatc1CreERT2;R26mTmG mice, 163 GFP+ cells and 1766 GFP- cells were quantified, respectively. For ProcrCreERT2-IRES-tdTomato;R26mTmG mice, 118 GFP+ cells and 1949 GFP- cells were quantified, respectively. n = 3 mice for each mouse model. Data represent the mean value ±SD ∗∗∗p < 0.001