Figure 4.
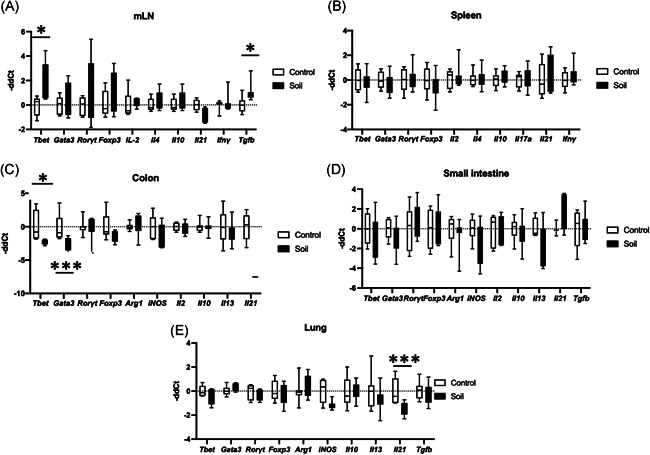
Gene expression changes in different organs after soil exposure. Mice were either treated with soil (n = 8) or left untreated (n = 8) as in Figure 1. RNA was then extracted from lymphoid organs (spleen and mesenteric lymph node [mLN]), intestine (small intestine and colon), and lung. Using quantitative polymerase chain reaction (qPCR) gene expression was measured. Open bars represent control animals and close black bars soil treated animals. T cell lineage‐associated genes (Tbet, Gata3, Rorɣt, and Foxp3), and cytokine expression (IL‐2, IL‐4, IL‐10, IL‐17a, IL‐21, and IFN‐ɣ) were measured in (A) mLN (Tgfβ was also analyzed for mLN) and (B) spleen. For the gastrointestinal organs, T cell lineage associated genes (Tbet, Gata3, Rorɣt, and Foxp3), macrophage activation associated genes (Arg1 and iNos) and cytokine expression (IL‐2, IL‐10, IL‐13, and IL‐21) were measured in (C) colon and (D) small intestine (Tgfβ was also analyzed for small intestine). Finally, T cell lineage associated genes (Tbet, Gata3, Rorɣt, and Foxp3), macrophage activation associated genes (Arg1 and iNos), and cytokine expression (IL‐10, IL‐13, IL‐21, and Tgfβ) were measured in (E) lung. Values are shown as box and whiskers (25–75 percentile). The data were analyzed with Mann–Whitney U test, *p < .05, **p < .01, ***p < .001
