Figure 5.
A pair of bilateral SEZ neurons, MooSEZs, underlies GH146II-GAL4 optogenetically induced backward locomotion
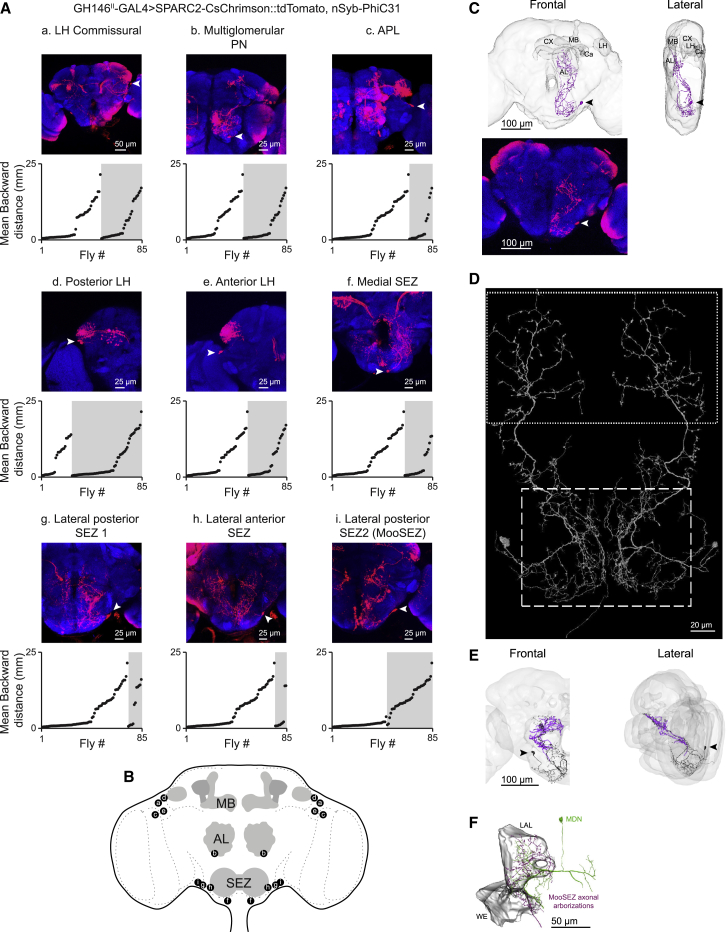
(A) GH146II-GAL4 expression pattern is composed of nine different neuronal clusters (a–i) in the central brain (as designated, arrowheads indicate somata). Maximum intensity projections of 24–111 confocal sections (1 μm) through the central brain are presented. The SPARC method was used to generate mosaic flies expressing CsChrimson::tdTomato in stochastically distributed subsets of neurons within the GH146II-GAL4 driver line. SPARC generated flies were individually exposed to a 2-s red light pulse in the open arena. For each of the identified neuronal cluster, the mean backward translations of individual flies during the light pulse averaged over three trials as a function of expression are presented. The gray shading indicates expression in the cluster of interest. Only for the MooSEZ cluster (lateral posterior SEZ 2) a clear lack of backward walking is observed when there is no expression and a clear backward locomotion when there is expression. Multiple linear regression analysis in which each cell cluster is set as a predictor of the mean backward translation indicates that MooSEZs exclusively correlate with backward locomotion (Table S2).
(B) Schematic drawing of the cell clusters identified in (A) using the SPARC genetic method.
(C) Top, three-dimensional light microscope reconstruction of a single MooSEZ. Frontal (left) and lateral (right) views are presented. The MooSEZ soma is labeled with a black arrowhead. MB, mushroom body; CX, central complex; AL, antennal lobe; Ca, calyx; LH, lateral horn. Bottom, expression pattern of a stochastically targeted MooSEZ expressing CsChrimson::tdTomato labeled by GH146II-GAL4 using the SPARC “Sparse” effector. MooSEZ morphology is faithfully recapitulated by the LM reconstructed MooSEZ presented on top. The MooSEZ soma is labeled with a white arrowhead. Maximum intensity projection of 69 confocal sections (1 μm) through the central brain is presented. For visualization purposes, the projection image is vertically flipped.
(D) Three-dimensional EM reconstruction of MooSEZ in both hemispheres identified in the FAFB using FlyWire. MooSEZ SEZ branches are thinner and show a higher degree of arborization typical of dendritic regions (dashed box), whereas protocerebral branches exhibit a higher probability of varicose and globular swellings, typical of presynaptic areas (stippled box).
(E) MooSEZ three-dimensional EM reconstructions in FlyWire (black), and in FlyEM hemibrain (purple). The partially reconstructed hemibrain MooSEZ highly resembles the fully reconstructed FlyWire MooSEZ within the hemibrain volume. Both EM reconstructions are similar to the fully reconstructed LM MooSEZ in (C). FlyWire MooSEZ soma is labeled with a black arrowhead. Frontal (left) and lateral (right) views are presented.
(F) Three-dimensional EM reconstructions of MooSEZ neurites (fragment #5813046185, purple) and a representative post synaptic MDN (fragment #5813021075, light green) identified in the FlyEM hemibrain using NeuPrint+. MooSEZ is synaptically coupled to MDNs. Frontal view of the lateral accessory lobe (LAL) and the wedge (WE) brain regions in the right brain hemisphere is presented.