Figure 7.
SMR4 decelerates the cell cycle via direct interactions with a selected set of D-type cyclins
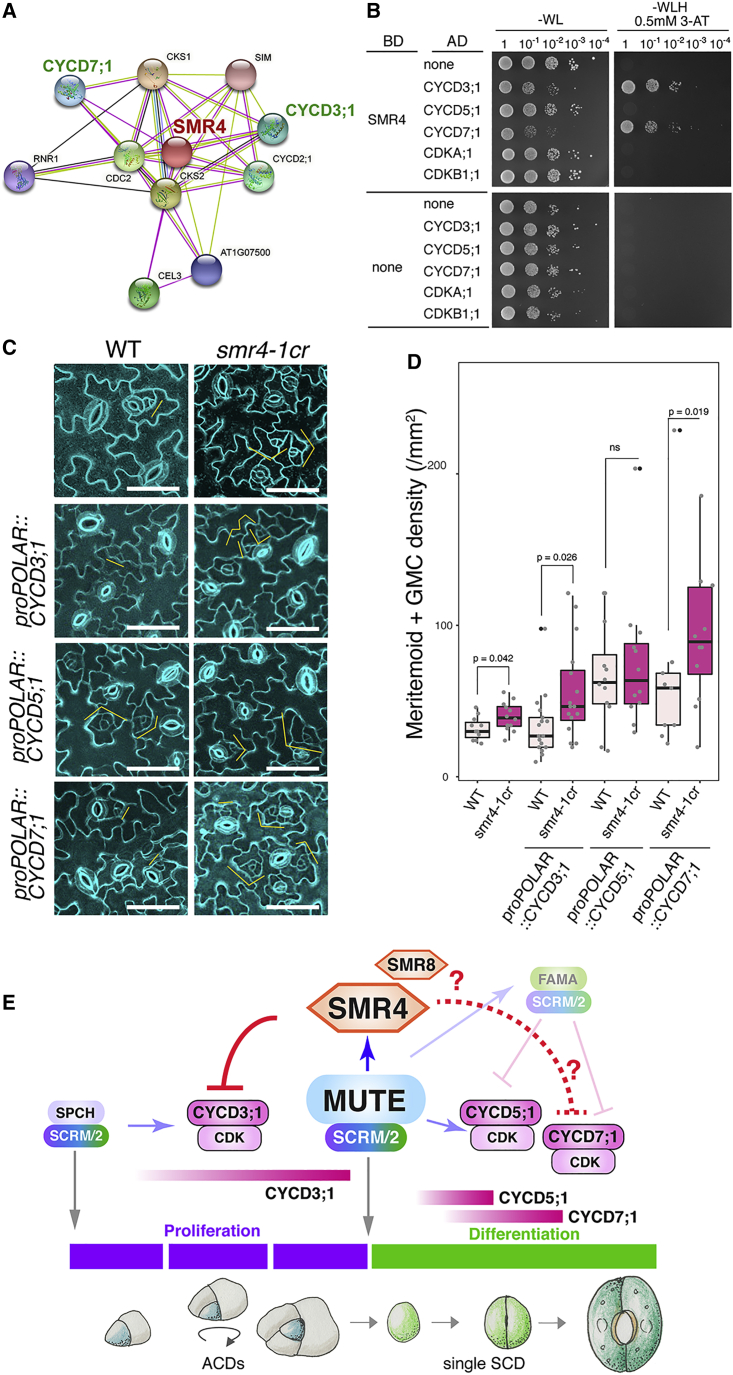
(A) SMR4 interacting proteins from in vivo interactome (Van Leene et al., 2010) visualized by cytoscape.
(B) Yeast two-hybrid assays. Bait, the DNA-binding domain (BD) alone or fused to SMR4. Prey, the activation domain alone (AD) or fused to CYCD3;1, CYCD5;1, CYCD7;1, CDKA;1, and CDKB1;1. Transformed yeast were spotted in 10-fold serial dilutions on appropriate selection media.
(C) Transgenic plants harboring CYCD3;1, CYCD5;1, and CYCD7;1 driven by the POLAR promoter in wild type (WT) and smr4-1cr in comparison with wild type and smr4-1cr. Orange brackets: stomatal lineage precursors. Scale bars, 50 μm.
(D) Quantification of stomatal precursor cells in 1.0 mm−2 area from 7-day-old seedlings. Mann-Whitney test was performed. p values were marked on top of the boxplot. Independent T1 transgenic plants were analyzed. The number of plants used: WT: n = 11, smr4-1cr: n = 12, proPOLAR::CYCD3;1: n = 17, proPOLAR::CYCD3;1 smr4-1cr: n = 16, proPOLAR::CYCD5;1: n = 12, proPOLAR::CYCD5;1 smr4-1cr: n = 12. proPOLAR::CYCD7;1: n = 9, proPOLAR::CYCD7;1 smr4-1cr: n = 12.
(E) Schematic model. SPCH·SCRM/2 initiate and sustain ACD and MUTE·SCRM/2 trigger SCD (gray arrows) by transcriptionally activating CYCD3;1 and CYCD5;1 (shaded blue arrows), respectively. MUTE directly up-regulates SMR4 transcription (Blue arrow). SMR4 (and SMR8 in part) suppress the activity of CYCD3;1 and possibly CYCD7;1 complexed with CDKs (red line), but not CYCD5;1, to terminate the ACD mode and ensure faithful progression of SCD. Question marks and dotted line indicate the possible roles of SMR8 in termination of ACD and SMR4 with CYCD7;1 in symmetric cell division, respectively.