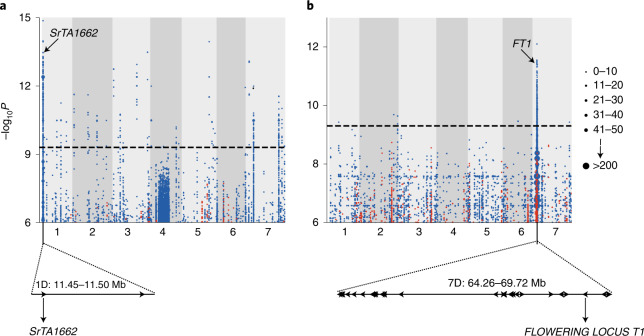
Fig. 2. Genetic identification of candidate genes for stem rust resistance and flowering time by k-mer-based association mapping.
a, k-mers significantly associated with resistance to Puccinia graminis f. sp. tritici race QTHJC mapped to scaffolds of a de novo assembly of Ae. tauschii accession TOWWC0112 anchored to chromosomes 1 to 7 of the D subgenome of Chinese Spring51. Points on the y axis show k-mers significantly associated with resistance (blue) and susceptibility (red). b, k-mers significantly associated with flowering time mapped to Ae. tauschii reference genome AL8/78 with early (red) or late (blue) flowering time association relative to the population mean across the diversity panel. Candidate genes for both phenotypes are highlighted. Point size is proportional to the number of k-mers (see inset). The association score is defined as the –log10 of the P value obtained using the likelihood ratio test for nested models. The threshold of significant association scores is adjusted for multiple comparisons using the Bonferroni method.