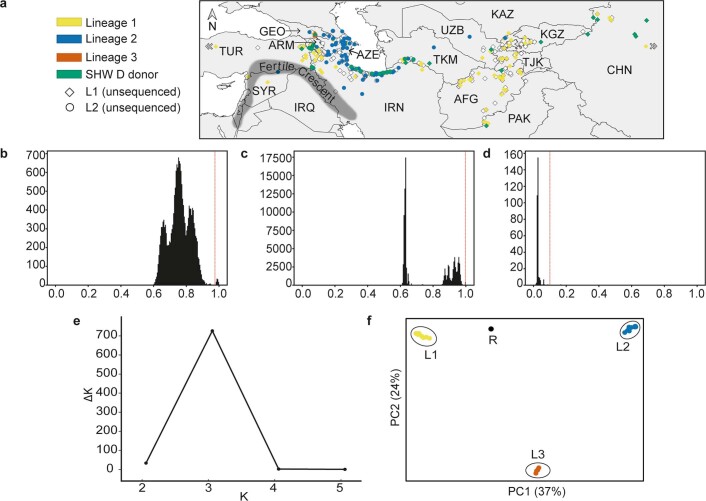
Extended Data Fig. 1. Configuration and genetic structure of the Aegilops tauschii diversity panel used in this study.
a, Geographical distribution of 242 Ae. tauschii accessions. Filled squares and circles represent accessions sequenced as part of this study, while accessions represented by unfilled squares and circles were not sequenced. Accessions highlighted in green were used as D genome donors to generate synthetic hexaploid wheat (SHW) lines. Three accessions outside of the map, one from Turkey and two from China, are indicated by white arrow heads. AFG, Afghanistan; ARM, Armenia; AZE, Azerbaijan; CHN, China; GEO, Georgia; IRN, Iran; IRQ, Iraq; KAZ, Kazakhstan; KGZ, Kyrgyzstan; PAK, Pakistan; SYR, Syria; TJK, Tajikistan; TUR, Turkey; TKM, Turkmenistan; UZB, Uzbekistan. The Fertile Crescent follows the shaded area in Fig. 1 of Harlan and Zohary (1966) and is bound by the Mediterranean in the west, by chains of large and high mountain ranges in the north and east (the Amanos in northwestern Syria, the Taurus in southern Turkey, Ararat in north-eastern Turkey and the Zagros in western Iran), and in the south by the Syrio-Arabian desert, with its western extension (for example, Paran desert) in the Sinai Peninsula. b, Identification of non-redundant Ae. tauschii accessions using KASP markers on 195 accessions and: c, 100,000 random SNPs obtained from whole genome shotgun sequencing of 306 accessions. The vertical red line in both histogram similarity plots indicates the redundancy cut-off at which the peak of the high similarity values is clearly separated from the rest. d, Identification of Ae. tauschii accessions with minimal residual heterogeneity. The histogram of heterozygosity scores was generated using all the bi-allelic SNPs obtained from whole genome shotgun sequencing of 305 accessions (excluding TOWWC0193). The vertical red line indicates the cut-off at which the cluster of the low heterozygosity values is clearly separated. e, ΔK plot for a STRUCTURE run with 10 randomly selected accessions each of L1 and L2 along with the five accessions of the putative L3 and the control L1-L2 RIL. f, Principal Component Analysis with the same set of accessions as used in panel a. The recombinant inbred control line is indicated by R.