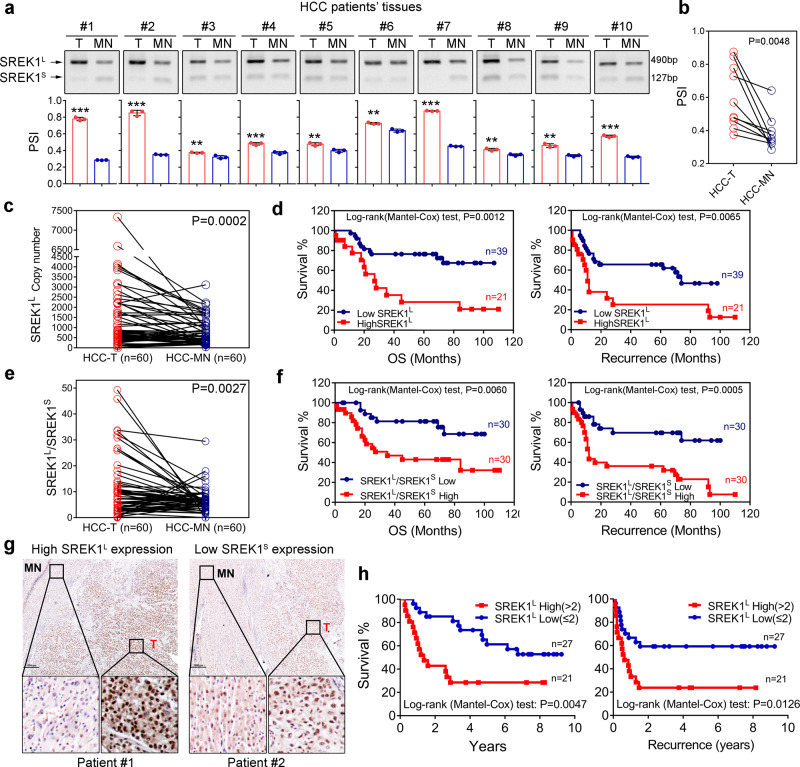
Fig. 1. The variant SREK1L is enriched in HCC and associated with prognosis.
PCR detection with primer set 1 and quantification of two variants in ten pairs of HCC patient tissues (Supplementary Table 1). Quantified relative PSI (Percentage-splice-in = splice_in/(splice_in+splice_out)) of the variants a in each tissues of ten pairs’ samples (n = 3, data are shown as the mean ± SD, two-tailed, unpaired t test is used) or b the average PSI (Percentage-splice-in = splice_in/(splice_in+splice_out)) in ten pairs tissues are shown (red column indicates tumor (T), Blue column indicates matched normal tissues (MN)), n = 10. c Normalized SREK1L copy number in 60 pairs of HCC tissues (Supplementary Table 2) and d survival analysis of 60 HCC patients with high and low expression of SREK1L (using the mean as the cut-off value; OS overall survival). e Normalized SREK1L/SREK1S expression ratios in 60 pairs of HCC tissues (Supplementary Table 2), and f survival analysis of 60 HCC patients with high or low SREK1L/SREK1S expression (using the median as the cut-off value). g Representative immunohistochemical (IHC) staining figures of high or low SREK1L expression in HCC tissues with SREK1L detection antibody, scale bar = 1000 μm. h Survival analysis on high or low SREK1L expression by IHC in 48 HCC patients was determined and scored independently by three pathologists. Two-tailed, paired t test is used for (b, c, e), **p < 0.01; ***p < 0.001. Source data are provided as a Source Data file.