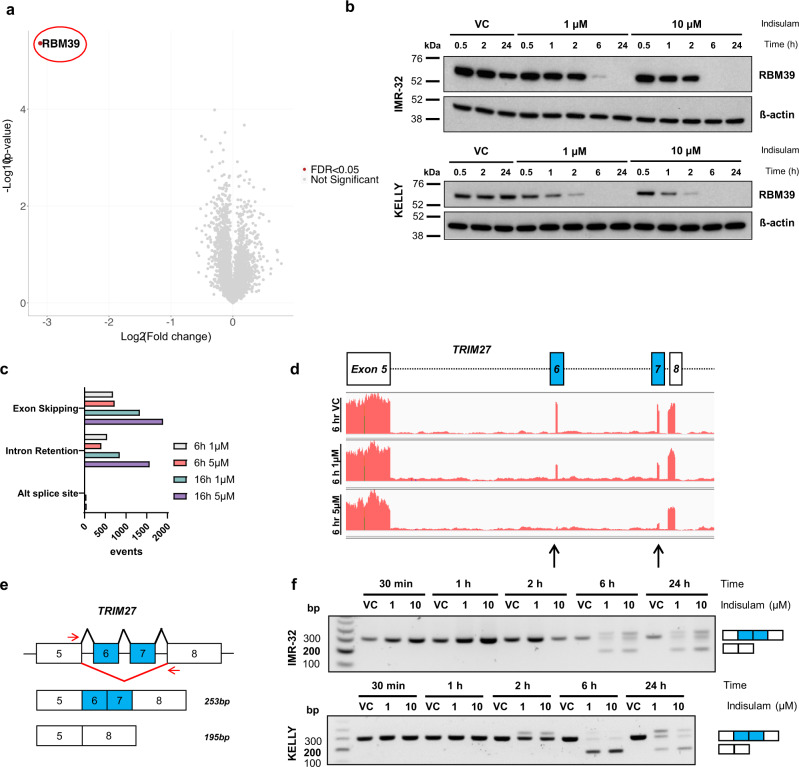
Fig. 2. Indisulam causes selective degradation of RBM39 and subsequent mis-splicing of RNA.
a Volcano plot of proteomic analysis in IMR-32 cells treated with 5 µM indisulam or VC (0.1% DMSO) for 6 h. RBM39 is highlighted in red. b Western blot of IMR-32 and KELLY cells treated with vehicle control (VC), 1 or 10 µM indisulam for 0.5–24 h. Representative blot for n = 2 independent experiments. c Number of RNA splicing events (SpliceFisher analysis, n = 3 independent experiments) in IMR-32 cells following treatment with VC or 5 µM indisulam for 6 or 16 h. d RNA read counts of TRIM27 (exon 5–8) in IMR-32 cells treated with VC (top), 1 µM (middle), or 5 µM (bottom) indisulam for 6 h. Black arrows indicate loss of exons 6 and 7. Plot generated with IGV. e Diagram of custom primers to detect skipping of exon 6 and 7 of TRIM27. f PCR of TRIM27 (exon 5–8) in IMR-32 (top) and KELLY (bottom) cells treated with VC, 1 or 10 µM indisulam for 0.5–24 h. Representative blot for n = 2 independent experiments. Source data is provided as a Source Data File.