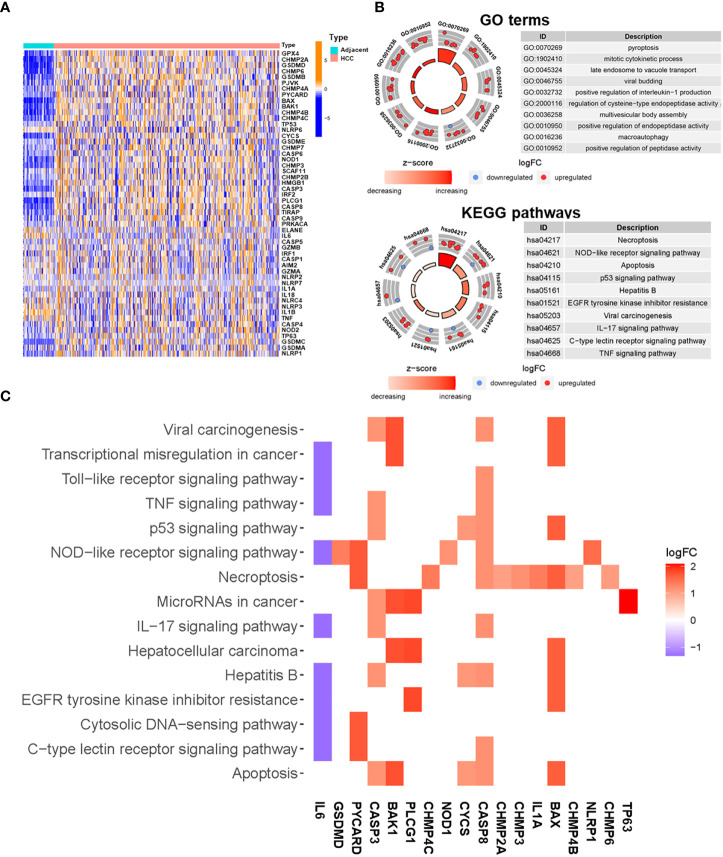
Figure 2.
Functional enrichment analysis of differentially expressed pyroptosis-related genes. (A) Heatmap of expression of pyroptosis-related genes in HCC and adjacent tissues (blue: low expression level; dark orange: high expression level). (B) Enriched GO terms (up) and KEGG pathways (down) for differentially expressed pyroptosis-related genes. The outer circle shows a scatter diagram of the log fold change (FC) value allocated to each term. (C) Heatmap showing the relationships between differentially expressed pyroptosis-related genes and enriched KEGG pathways. Colors represent the logFC values of each gene in HCC compared with adjacent tissues.