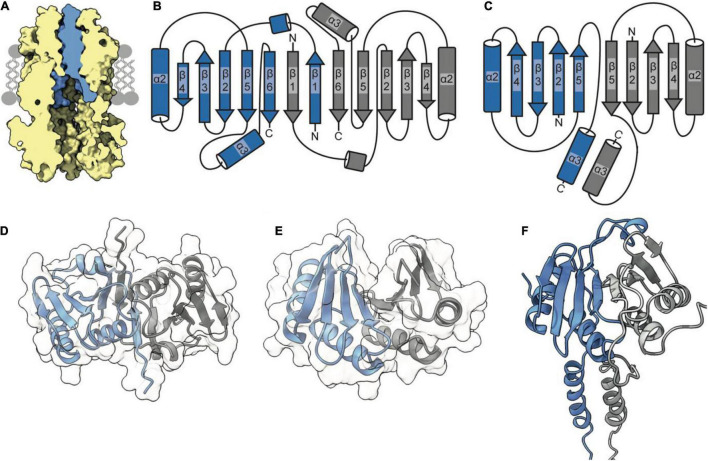
FIGURE 2.
The TolR dimer has two distinct conformations. (A) The periplasmic entrance to the TolQ (yellow) pore is blocked by the transpore helices of the TolR dimer (blue). The TolQ-TolR model shown was assembled following alignment of the AlphaFold TolR monomer with the truncated TolR dimer from H. influenzae (TolRH). The full-length TolR dimer was then docked into the TolQ pentamer previously generated (Figure 1) based on position of ExbD in the ExbBD complex structure (Celia et al., 2019). The final TolQ-TolR complex shown was determined following energy minimization using RosettaRelax. (B) A depiction of the secondary structure of TolR from E. coli (TolRE) highlighting the strand-swapped dimer formed by β1 and β6 of each monomer. (C) A depiction of the secondary structure of TolRH has a dimer interface formed through interactions of β5-β5. (D) The TolRE strand-swapped dimer (PDB ID 5BY4) solved by X-ray crystallography shows the 180° rotation of one monomer (gray) relative to the other (blue). (E) The TolRH structure solved by NMR (PDB ID 2JWK) is in an open PG-binding conformation. Each monomer is represented in blue and gray, respectively. (F) The E. coli TolR dimer, generated in Rosetta, models the entirety of the protein, which adopts an open conformation like that of TolRH.