Abstract
The mitotic count (MC) is an important histological parameter for prognostication of malignant neoplasms. However, it has inter- and intraobserver discrepancies due to difficulties in selecting the region of interest (MC-ROI) and in identifying or classifying mitotic figures (MFs). Recent progress in the field of artificial intelligence has allowed the development of high-performance algorithms that may improve standardization of the MC. As algorithmic predictions are not flawless, computer-assisted review by pathologists may ensure reliability. In the present study, we compared partial (MC-ROI preselection) and full (additional visualization of MF candidates and display of algorithmic confidence values) computer-assisted MC analysis to the routine (unaided) MC analysis by 23 pathologists for whole-slide images of 50 canine cutaneous mast cell tumors (ccMCTs). Algorithmic predictions aimed to assist pathologists in detecting mitotic hotspot locations, reducing omission of MFs, and improving classification against imposters. The interobserver consistency for the MC significantly increased with computer assistance (interobserver correlation coefficient, ICC = 0.92) compared to the unaided approach (ICC = 0.70). Classification into prognostic stratifications had a higher accuracy with computer assistance. The algorithmically preselected hotspot MC-ROIs had a consistently higher MCs than the manually selected MC-ROIs. Compared to a ground truth (developed with immunohistochemistry for phosphohistone H3), pathologist performance in detecting individual MF was augmented when using computer assistance (F1-score of 0.68 increased to 0.79) with a reduction in false negatives by 38%. The results of this study demonstrate that computer assistance may lead to more reproducible and accurate MCs in ccMCTs.
Keywords: canine cutaneous mast cell tumors, artificial intelligence, digital pathology, deep learning, mitotic figures, mitotic count, automated image analysis, computer assistance
Proliferation parameters of neoplastic cells correlate with patient prognosis for many tumor types, including canine cutaneous mast cell tumors (ccMCTs), and are a relevant criterion for treatment recommendations that come with considerable financial and quality of life implications. 6,9,24,37,42 The mitotic count (MC) is the only proliferation marker that can be determined quickly and efficiently in standard histological sections stained with hematoxylin and eosin (HE) and is therefore routinely evaluated in every potentially aggressive tumor type. 28,37 For ccMCT, the MC has been used either as a solitary parameter 9,35,41 or as part of tumor grading systems. 27 If used as a solitary prognostic parameter, a 2-tiered system with a MC of 0 to 5 and a MC >5 has been evaluated to yield a sensitivity of 32%, 39%, 50%, and 55% (high percentage of false negatives) and specificity of 91%, 96%, 98%, and 99% (low percentage of false positives) regarding ccMCT-related deaths in different studies. 8,9,41,42 In order to increase sensitivity, other research groups have proposed using a cutoff value of MC ≥ 2 (sensitivity: 76% and 84%; specificity: 56% and 80% for ccMCT-related death) 24,41 or to use a stratification of the MC into 3 groups (0, 1–7, >7 and 0–1, 2–7, >7, respectively; sensitivity and specificity not available). 19,41 These data prove that the MC is relevant for prognostication of ccMCT; however, it also reveals some variability in the derived results, which causes uncertainty for routine evaluation of mitotic density and interpretation of its prognostic value. The question arises whether this variability could be reduced with standardization of the MC methods between different studies that would ultimately allow more appropriate treatment recommendations.
The MC is usually defined as the number of mitotic figures (MFs, or cells undergoing mitosis visible with microscopy 18 ) within “10 high-power fields (HPFs)” with the highest mitotic density, that is, hotspot tumor location. 18,31,32 Looking at this definition, 3 potential sources of variability for performing the MC become apparent: (1) variable size of the evaluated tumor area (“10 HPFs”), (2) selection of different tumor areas in cases with variable distribution of mitotic figures (mitotic density) throughout the tumor section, and (3) inconsistent identification of individual MFs at high magnification and classification against imposters. 18,31
Some studies have proven that it is feasible to perform the MC using digital microscopy and whole-slide images (WSIs). 1,7,13,38,46 As opposed to light microscopy, the concept of enumerating 10 HPFs (round fields using a microscope at 400× magnification) is extraneous as the area can be accurately measured and labeled in the WSI (with a rectangular field of view). In light microscopy, the size of a single field of view at 400× magnification may vary notably depending on the field number of the microscope, and a standard size of 2.37 mm2 (based on the field number of 22) has been proposed for veterinary pathology. 31,32 Instead of using the term “10 HPFs” for this study (that used WSI), we use the term “mitotic count region of interest” (MC-ROI) for a single, rectangular area with the size of 2.37 mm2 in the tumor location with the presumed highest mitotic density. 5,11
Regarding the selection of the MC-ROI, it is common practice to attempt to find a single tumor area with the highest mitotic activity, that is, a “hotspot.” 8,27,31,32,35 It has often been assumed (but rarely proven, eg, in human breast cancer 25 ) that the most mitotically active tumor area correlates best with biological behavior of the neoplasm. Of note, it has been shown that the mitotic density varies notably between different tumor locations in histological sections of ccMCTs and canine mammary carcinomas 3,5,11 and that pathologists have some difficulties in finding hotspots. 3,11,46
The pathologist’s capability to identify and classify MFs has been evaluated recently. 16,44,45,47 Those studies compared the digital MCs of different pathologists in the same MC-ROIs and revealed an overall difference in number of annotated MFs by a factor of 1.5× to 3.3×. 45,47 This can be attributed to erroneous MF detections related to failure in identification of MF candidates, as well as to inaccurate or inconsistent classification of MFs against lookalikes (such as apoptotic bodies, hyperchromatic or deformed nuclei, and inflammatory cells). 18 Tabata et al 38 have shown that pathologists have a lower accuracy of MF detection when using WSIs (69% to 74%) as compared to traditional light microscopy (80%).
Despite the potential limitations of digital microscopy in detecting MFs, WSIs enable innovative computerized image analysis approaches that have the potential to improve MC reproducibility and accuracy and thereby may improve standardization of the MC methods. 10,14 Development of high-performance image analysis algorithms for MFs has only been possible in the last decade due to the availability of groundbreaking machine learning solutions (especially deep learning using convolutional neural networks) and availability of large-scale datasets. 3,12,43,45 Regardless of these promising advancements, one of the main points of criticism of deep learning is its “black box” character (ie, the unavailability of decision criteria) that may result in failure of identifying algorithmic failure. 10,30 In order to ensure high reliability, approaches that allow review of the algorithmic predictions by trained pathologists (computer-assisted diagnosis/prognosis) through visualization of algorithmic results as an overlay on the WSI have been recommended for future application. 10 First computer-assisted MC software solutions have been recently validated for human pathologists. 7,33 However, studies that validate the utility of computer assistance for each critical step of performing the MC (see above) and for veterinary tumor histopathology have not been published to date.
The aim of the present study was to compare partially (MC-ROI preselection) and fully (additional MF candidate proposal) computer-assisted MC analysis with routine (unaided) MC analysis in WSIs of ccMCT. We evaluated the assistive value and limitations of computer assistance on the ability of 23 pathologists to perform the overall MC, to determine a count below or above a prognostic cutoff, to select a hotspot MC-ROI, and to identify and classify individual MFs. The ultimate goal was to identify a computer-assisted approach that may be helpful for standardization of the MC.
Material and Methods
Course of Studies
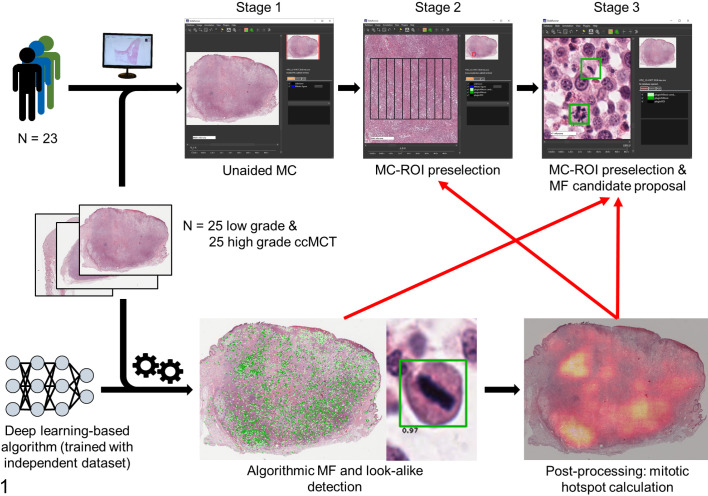
For this study, anatomic pathologists (study participants) performed MCs in WSIs of 50 ccMCTs at 3 stages with different degrees of computer assistance (none, partial, and full; Fig. 1). In stage 1, there was no computer assistance and participants were tasked with screening the WSI manually for MC-ROIs (mitotic hotspots) and annotating all MFs (including atypical MFs) within this area using their “routine” approaches. For stage 2 (partial computer assistance) and stage 3 (strong computer assistance), WSIs were analyzed with a deep learning–based algorithm that detected MFs in the entire tissue section. Based on the algorithmic MF detections, the MC was calculated for each possible tumor location resulting in the MC distribution. The ROI with the highest MC was preselected automatically and presented to the participants in stage 2. For stage 3 (full computer assistance), in addition to the same algorithmic MC-ROI preselection as used in stage 2, visualization of the individual MFs and MF lookalike detections (to assist MF identification) were provided as an overlay on the WSI along with their corresponding algorithmic confidence values (to assist MF classification). These algorithmic detections were only used as an aid to identify and classify potential mitotic figures, and participants had to annotate each structure that they wanted to count as a mitotic figure.
Figure 1.
Overview of the course of the study (stages 1–3) with different degrees of computer assistance (red arrows) of the study participants in 3 examination time points (stages). The deep learning model (concatenation of 2 convolutional neural networks) that was used in this study for computer-assistance was developed in a previous study using an independent training and test dataset with different WSI from the same laboratory that provided the study cases. 12 ccMCT, canine cutaneous mast cell tumors; MC-ROI, mitotic count region of interest; MF, mitotic figure.
Participants were instructed to follow the course of the 3 stages strictly and to wait at least 3 days until the next stage (often, there were multiple weeks between 2 stages). While performing the MCs, participants labeled the enumerated MFs (including atypical MFs) in the exact location of the digital image with a specialized annotation software. This method allows determination of the ability of participants and the ability of the deep learning–based model to identify individual MFs (on the object-level) compared to a gold standard–derived (pHH3 IHC-assisted) ground truth dataset. After the study, participants were asked to fill out an opinion survey.
Study Cases
Random ccMCT cases with an equal distribution of low and high histologic grade 27 (based on the original pathology reports) were selected from the archive of the Institute of Veterinary Pathology, Freie Universität Berlin. One tissue block from each of these cases with the largest tumor area was selected. Histological sections were produced from each of the blocks and stained with HE using the same tissue stainer (ST5010 Autostainer XL; Leica) at different batches and time points. Glass slides were digitized with a linear scanner (ScanScope CS2; Leica) using default settings. WSIs with one focal scan plane were produced at a magnification of 400× (image resolution: 0.25 µm per pixel). Specimens with overall very poor tissue preservation (ie, marked loss of nuclear details in most of the tumor section) and cases with a tumor section of <12 mm2 (measured by polygon annotation) were excluded. This procedure was followed until 35 low-grade cases and 35 high-grade cases (based on the original pathology reports) were selected. Clinical follow-up (patient outcome) was not considered for this study as the main goal was validation of different MC methods and not to determine prognosis. Local authorities (State Office of Health and Social Affairs of Berlin) approved the use of the samples (approval ID: StN 011/20) for research.
The 70 cases were analyzed with a deep learning–based MF algorithm (see below). Of these, 7 cases (all low grade according to the original reports) had a computerized MC-ROI preselection outside of the tumor area (due to MF predictions mainly in the epidermis, hair follicles, reserve cells of the sebaceous glands, or areas of crush artifacts) and were excluded from the study. The remaining 63 cases contained algorithmic MC-ROI preselection within the tumor area and were not further evaluated for exclusion purposes. We randomly excluded 3 additional low-grade and 10 high-grade cases (according to the original reports) in order to reduce the study set to 25 low-grade and 25 high-grade ccMCTs. Cases of the study set were randomly assigned numbers from 1 to 50. Additionally, a test slide (high-grade ccMCT) was provided to allow familiarization of study participants with the annotation software, the study tasks, and the properties of the digital images.
Study Instructions to Participants
Twenty-six pathologists from 13 different laboratories volunteered to participate in this study. The study material (WSIs, annotation software, files with algorithmic predictions) was provided, the goal and course of the study was explained, and the annotation software was demonstrated (using the test slide) to each participant. For stage 1, participants were instructed to search for mitotic hotspot MC-ROIs. However, no specific recommendations were given on how to find the “correct” MC-ROI. For all 3 stages, participants were instructed to annotate all MFs in the MC-ROIs with high diligence using their “routine” decision criteria. No specific diagnostic criteria for “correct” identification and classification of MFs (including atypical MFs) were provided in order to validate a realistic diagnostic setting.
Annotation Software and Database Creation
The open source software SlideRunner 2 was used in this study for annotating each enumerated MF in the precise pixel position in the digital image (object-level). Beyond the ability to view and navigate WSI, this software includes tools for fast image annotations, which are automatically stored in a database. For this study, each participant created their own databases (one for each stage) and annotated each MF present in the MC-ROIs using a single click tool (saving the x and y coordinate in the WSI). Participants were instructed to pay close attention in placing the annotation in the center of the MFs.
SlideRunner allows integration of software plugins for computerized image analysis and visualization of algorithmic predictions. During specimen selection, all WSIs were analyzed by an MF algorithm plugin (see below), and predictions of the study cases were saved as separate files that were provided to the participants for visualization in stages 2 and 3.
For each study stage, a different SlideRunner package was created, each including a different plugin that enabled visualization of the computerized information relevant for the respective stage as an overlay on the WSI. As intended by the study design, the stage 1 package of the software did not allow visualization of any algorithmic detections. However, this package included a plugin for a rectangular box with the size of exactly 2.37 mm2 (aspect ratio of 4:3, width of 1777.6 µm, and height of 1333.2 µm). The box was divided into a grid with 9 vertical lines (the distance between lines was less than the width of the field of view at 400× magnification), which intended to improve navigation in a meandering pattern. Participants were able to move the box to the desired MC-ROI location by re-centering the box coordinates to the present field of view at any viewing magnification. Due to a software failure, the exact image location of the selected MC-ROIs was not saved in the database. Therefore, we retrospectively determined the approximate MC-ROIs in which the highest number of annotations in the image could be placed. This ensured that the shift between the approximate and the actual selected MC-ROIs were minimal and negligible for our analysis (see below). For 63 instances that did not have an MF annotation (pathologist’s MC = 0), an approximate MC-ROI could not be determined retrospectively for the respective participant and WSI.
The plug-ins of the SlideRunner packages of the 2 subsequent stages visualized the predictions of the deep learning–based algorithm (Fig. 1). Stage 2 included a plugin with a 2.37 mm2-sized rectangular box (as described above) placed in the region that had the highest density of algorithmic MF detections (partial computer assistance, Suppl. Fig. S1). Unlike stage 1, participants were unable to move the box to another tumor location, even if they considered this not to be the most appropriate tumor location. This was done in order to enable comparison of the pathologist’s performance to identify and classify individual mitotic figures in the same tumor locations. The software package for stage 3 displayed the fixed 2.37 mm2 box in the same tumor area and additionally visualized all predicted MFs (as dark green boxes) and predicted MF lookalikes (as light green boxes) along with their algorithmic classification scores (“confidence value”; Suppl. Fig. S2). We decided to visualize the MF lookalikes in order to account for potential false-negative algorithmic predictions. Algorithmic predictions were only intended as an aid to find potential candidates and classify them as mitotic figures, but pathologists still had to annotate each structure that they wanted to count as a MF or disregard the algorithmic detection if they did not want to count this structure. Pathologists were also instructed to annotate MFs that were not detected by the algorithm.
After participants finished stage 3, they submitted the 3 databases to the principal investigators, who verified that all cases had been examined. All annotations that had a center coordinate outside of the 2.37 mm2 boxes were deleted as those represented erroneous or unintentional annotations. The participants were anonymized by assigning a random identification number to each participant.
Image Analysis Algorithm
Predictions of the deep learning–based algorithm were used for stages 2 and 3. Computerized image analysis comprised 2 analysis tasks as previously described by Aubreville et al: 5 (1) a deep learning model that detects MFs and (2) postprocessing steps composed of computerized MC density calculation (heat map) and hotspot MC-ROI preselection.
Briefly, the detection of MFs is based on a concatenation of 2 convolutional neural networks (RetinaNet and ResNet18 architecture). 5 The first convolutional neural network (object detector) was used to screen the entire WSI for potential MF candidates with high sensitivity and a high processing speed. The second convolutional neural network (patch classifier) was developed to classify small image patches of the detected candidates (by the first neuronal network) into MFs (classification threshold ≥0.5) and MF lookalikes (classification threshold <0.5) with high specificity. The model classification scores (“confidence value”) were extracted from the patch classifier (in order to display along with the algorithmic predictions) and ranged between 0.01 (very unlikely to be a MF) and 1.0 (very likely to be a MF). The models were trained and technically evaluated with an open access dataset with 44 880 MF annotations in 32 ccMCT WSIs that were produced by the same institute that provided the cases for the present study (using the same staining protocol and WSI scanner). 12 The ground truth for this training dataset of the algorithm was created by 2 pathologists (the principle investigator and a participant of this study) via consensus of each label using HE images only (pHH3 immunolabeling was not available).
Predictions of the concatenated convolutional neural networks were used to derive the MC density map by computerized calculation of the MC, that is, the number of algorithmic MF detections within a 2.37 mm2 box (see above), for every possible center coordinate of a 2.37 mm2 box that contains more than 95% tissue. 5,11 The MC-ROIs for stages 2 and 3 were selected as the image location with the highest MF density in the MC map.
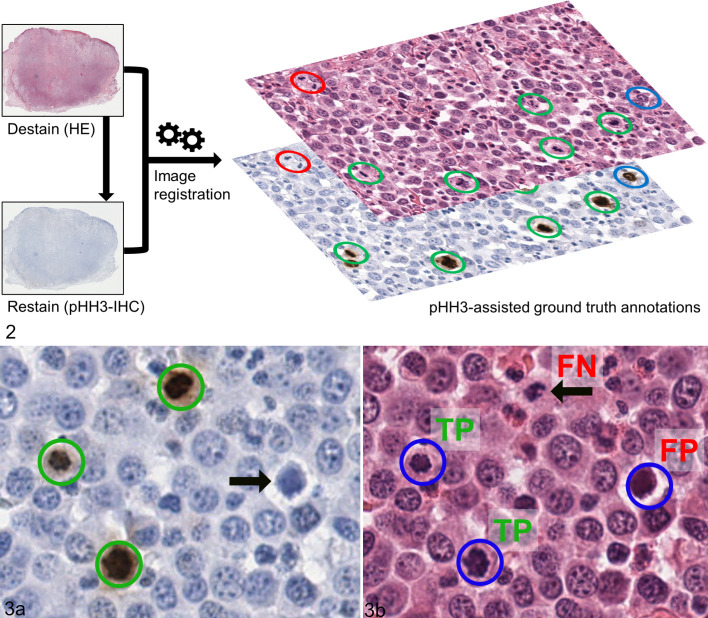
pHH3-Assisted Ground Truth
In order to evaluate the pathologists and algorithmic performance on the object level (identify and classify individual MFs in the MC-ROIs at high magnification), a ground truth dataset was developed with the assistance of immunohistochemical labeling for phosphohistone H3 (pHH3). pHH3 is a DNA-binding protein that is mostly specific for the mitotic phase of the cell cycle. Histone H3 is phosphorylated in early prophase (still indistinct in HE sections) but already dephosphorylated in telophase. 23 Based on Tellez et al, 39 we established a protocol to destain the initial HE-stained sections and immunohistochemically label the same tissue section in order to ensure that the exact same cellular objects were represented in both WSIs. The coverslips of HE-stained sections were removed by incubation in xylene. Subsequent to incubation in a descending alcohol series (99%, 80%) slides were destained under visual control in a 70% alcohol solution with 0.37% hydrogen chloride. After destaining, immunohistochemistry was performed, including blockage of endogenous peroxidase (with 10% H2O2), antigen retrieval by microwave heating (with citrate buffer), and antigens blocked (with goat serum). For the primary antibody, we used Phh3 clone E173 (rabbit monoclonal, ab32107, Abcam), as this product was used in a previous canine study. 34 Because the HE-stained sections of the study cases were produced at least 1 year earlier, a higher antibody concentration of 1:650 (as opposed to new tissue sections with 1:1500) was necessary (established on the excluded cases from this study; see above). The secondary antibody was a goat anti-mouse IgG (H+L) conjugated with alkaline phosphatase and incubated at a dilution of 1:200. 3,3′-Diaminobenzidin (DAB) was used as a chromogen and hematoxylin as counterstain. ccMCTs were used as positive and negative controls. Immunolabeled glass slides were digitized as described above. In 10 of the 50 tissue sections included in the study, relevant tissue parts were lost during immunohistochemical processing (if not mounted on adhesive glass slides) or immunohistochemical labeling was nonspecific (internal control compared to the HE staining). Therefore, those cases were excluded from the pHH3-assisted labeling portion of this study and were not available for performance evaluation on the object level.
Following this procedure, we produced 2 WSIs (one stained with HE and the other labeled with pHH3) from the same tissue section for 40 of the 50 cases. In contrast to using recut tissue sections, this process ensured visualization of the same cellular objects in both WSIs. Automated image registration (according to Jiang et al 26 ) was performed in order to align the 2 images so that the tissues matched almost perfectly on a cellular level. Using a newly developed SlideRunner plug-in, it was possible to instantly switch between images with the 2 staining methods and therefore easily compare the information for each cell (Fig. 2). The principal investigator (a pathologist not involved as a study participant) developed a ground truth dataset (pHH3-assisted ground truth) for the algorithmically selected MC-ROIs of stages 2 and 3 in the HE images by annotating all the cells that were positive for pHH3 and additionally annotated the unambiguous late phase MFs that failed to label for pHH3 (late phase MFs were a small proportion of all annotations). In addition, a pHH3-assisted count was performed in the manually selected MC-ROIs (stage 1) if the MC of the respective participant was higher than in stage 2 in order to evaluate which hotspot MC-ROI (manually or algorithmically selected) had a higher mitotic density (based on the pHH3-assisted count).
Figures 2–3.
Immunohistochemistry-assisted ground truth. Figure 2. Labeling method of the ground truth dataset. The histological sections (hematoxylin and eosin stain, HE) were destained and relabeled with immunohistochemistry against phosphohistone H3 (pHH3). Subsequently, whole-slide images of both staining methods were aligned on the cellular level via automated image registration and combined to decide if a tumor cell has a mitotic figure (MF) or not. Ground truth annotations comprised pHH3-positive cells that were recognizable on HE images (green circles) or were not readily identifiable on HE images (blue circles; especially prophase MF) as well as unambiguous late phase (especially telophase) MF that were pHH3-negative (red circles). Here these patterns are displayed as 3 distinct colors but in the ground truth dataset those structures were labeled as one label class. Figure 3a. High-magnification image of a pHH3-stained tumor section used for creating the ground truth with 3 positive tumor cells (green circles) and a pHH3-negative mitotic figure imposter (arrow). Figure 3b. Histological image (HE stain) of the same tumor location as in Figure 3a with exemplary annotations by one of the study participants (blue circles). Compared to the pHH3-assisted ground truth, 2 annotations are true positives (TP), 1 annotation is a false positive (FP), and 1 annotation MF was missed (false negative, FN, arrow).
Performance Evaluation
The MC was defined as the number of annotations by participants within a MC-ROI. The pHH3-assisted MC was the number of ground truth annotations in the respective MC-ROI. Interobserver agreement of the MCs between the 3 stages was calculated by the interobserver correlation coefficient (ICC) and its 95% confidence interval (95% CI). The ICC describes how strongly the pathologist’s MC values of the same tumor case resemble each other. ICC were evaluated as poor = 0 to 0.39, fair = 0.40 to 0.59, good = 0.6 to 0.74, and excellent = 0.75 to 1.00. 11 Differences were considered significant if the 95% CI did not overlap. 21 The coefficient of variation (CV) between the pathologists at each stage was calculated. The CV (in percent) is defined as the ratio of the standard deviation to the mean. Smaller CV percentage values represent lower variability.
Performance of classifying MCs into low and high according to the prognostic cutoff of MC ≥ 5 8,35,42 was measured by accuracy as the number of correctly (below or above cutoff according to the pHH3-assisted ground truth) classified instances divided by all instances. To calculate the P value for the difference in accuracy between the 3 stages, a generalized linear mixed model (GLMM) was used. We fitted a logistic regression using correct classification (1 = correct, 0 = incorrect) as outcome and using a random effect for the pathologist to account for repeated measures (40 slides per pathologist).
For comparison of the manually selected approximate MC-ROIs, we produced images that visualize the region and a MC heatmap as an overlay on the WSI. The MC heatmap was based on the unverified algorithmic predictions and was calculated as previously described. 11
The performance of the participants and algorithm to identify and classify individual MFs in the MC-ROIs of stages 2 and 3 (object detection task) was determined by standard object detection metrics that are commonly used for evaluation of MF algorithms. 3,10,12,16,33,43 –45,47 True positives (TP), false positives (FP), and false negatives (FN) were calculated against the pHH3-assisted ground truth (Fig. 3). A pathologist’s annotation and a ground truth annotation were counted as TP if the center coordinates of both had a maximum Euclidean distance of 25 pixels (equivalent to 6.25 µm). True negatives are not available for object detection tasks. 10 Precision (also known as positive predictive value, evaluates how many of the annotated cells are “true” MF according to the ground truth), recall (also known as sensitivity, evaluates how many of the ground truth MF were annotated by the participants) and the F1-score (harmonic mean of precision and recall) were defined as described by Bertram et al 10 : precision = TP/(TP + FP); Recall= TP/(TP + FN); F1 = 2× (precision × recall)/(precision + recall). Macro-averaged values were determined with the overall TP, FP, FN annotations or predictions of all cases totaled, and thus every MF identification and classification had the same weight regardless of the mitotic density of the individual images. This allows evaluation of the general performance of detecting each individual MF. 48 For the micro-averaged values, we determined the metrics (precision, recall, F1-score) for each slide and participant, and subsequently calculated mean of these values. 48 With this method, every image has the same weight, and the metrics represent the diagnostic performance on the sample level (individual MFs of cases with low mitotic density have a higher weight than individual MFs of cases with high mitotic density). This reflects more the diagnostic situation, where it is essential to have a particularly high performance on the object level in low and borderline mitotic density cases. In cases with an extremely high number of MFs (much higher than the prognostic MC cutoff), pathologists might experience a higher degree of fatigue or search satisfaction bias, which has a lower impact on the micro-averaged values. The 95% CI for the micro-averaged precision, recall, and F1-score were determined using bootstrapping (5000 replicates, bias-corrected and accelerated CI). Differences between the object detection metrics were considered significant if the 95% CI did not overlap. 21 The 95% CI values cannot be calculated for macro-averaged values.
A recall bias between the 3 stages was considered small for multiple reasons: manual selection of the MC-ROI was only done in stage 1 (but not in stages 2 and 3), the tumor region evaluated at high magnification was almost always different between stage 1 and stages 2 and 3 (thus different MFs were evaluated), the tumor regions evaluated twice (in stages 2 and 3) contained 1263 to 4807 MFs per stage (according to the participants annotations, see Results section), participants had to annotate each individual mitotic figure (as opposed to give a single diagnosis per case) and distinguish these from other structures (including lookalikes) in these images, and the amount of computerized information continuously increased from stages 1 to 3.
Results
Twenty-three of 26 (88%) pathologists from 11 laboratories completed this study. The remaining pathologists were excluded from analysis as they did not return results (N = 2) or did not examine all cases (N = 1). Of the included participants, 22 (96%) were Diplomates of the American (N = 12) or European (N = 10) College of Veterinary Pathologists and one (4%) had completed a residency in veterinary anatomic pathology. The duration of board certification ranged from 0 to 14 years (median of 6 years). Experience of the participants with performing MCs and digital microscopy is listed in Supplemental Tables S1 to S3 (experience level may not be representative for some laboratories). Dogs of the included cases had a median age of 9 years (range: 4–13 years). Breeds of the included cases are listed in Supplemental Table S4. Time to automatically analyze the study WSI by the deep learning–based algorithm (inference time) was on average 3:51 minutes (median time: 3:47 minutes; range: 1:41–7:13 minutes) using a PC running Linux with Intel Xeon E5-1630 CPU (4 cores @ 3.7 GHz), NVIDIA GTX 1080 graphics processing unit and SATA 3.0-connected SSD. This inference time includes analysis by the deep learning model (detection of MFs and classifying their confidence values), and the subsequent postprocessing step (heat map calculation), which took only a few seconds.
All 23 participants examined the 50 cases at the 3 examination stages (3450 pathologist evaluations) and thereby created 38 491, 59 634, and 68 570 annotations in stages 1, 2, and 3, respectively. The number of annotations (all 50 cases combined) per participant ranged between 549 and 4182 (mean: 1674; CV: 43%) for stage 1, between 1263 and 4412 (mean: 2593; CV: 33%) for stage 2, and between 1827 and 4807 (mean: 2981; CV: 21%) for stage 3. The pHH3-assisted ground truth dataset comprised 2617 annotations (40 cases only) and the deep learning–based algorithm predictions comprised 3063 MF candidates (50 cases, without review by participants) for the ROIs of stages 2 and 3.
Computer-Assisted MCs Have Higher Agreement
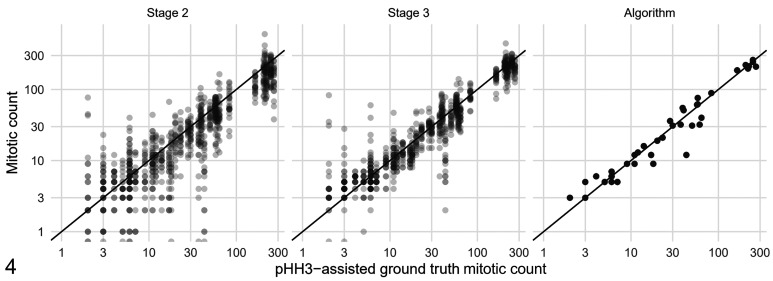
First, we evaluated the effect of computer assistance on the overall MC. The average MC for all cases and pathologists was 33.47 for stage 1, 51.86 for stage 2, 59.63 for stage 3, and 61.26 for the deep learning–based algorithm. Compared to stage 1, the average pathologist MC was increased by 54.9% in stage 2 and 78.2% in stage 3. Number of cases in which the MCs had very low values were notably reduced in stages 2 and 3. For example, MC = 0 were determined in 63 instances in stage 1 and only in 12 and 3 instances in stages 2 and 3, respectively (see Suppl. Fig. S3 for more MC values). Agreement of the MCs was higher between stages 2 and 3 (examination of the same MC-ROI) than agreement between stages 1 and 2 (mostly different MC-ROI; Suppl. Fig. S4). Interobserver agreement of the MCs was good for stage 1 (ICC: 0.70; 95% CI: 0.60–0.79) and excellent for stage 2 (ICC: 0.81; 95% CI: 0.74–0.88) and stage 3 (ICC: 0.92; 95% CI: 0.88–0.96). The 95% CI of the ICC for stage 1 and stage 3 do not overlap; thus, the difference was considered statistically significant. The CV for the MCs was 78.2% for stage 1, 51.3% for stage 2, and 34.9% for stage 3, thus reduced by more than half with full computer assistance. Figure 4 shows the improvement in agreement of the MCs with the pHH3-assisted ground truth MCs if computer assistance for identification and classification of MFs (stage 3 as opposed to stage 2) was available.
Figure 4.
Scatterplots of the participant’s mitotic count (MC) values (stages 2 and 3) and the algorithmic (unverified) MC compared with the pHH3-assisted ground truth MC (all obtained in the same mitotic hotspot MC-ROI based on the algorithmic heatmap). The black line in the scatterplots indicate equal values for ground truth and pathologists or algorithmic MCs.
Computer Assistance Improves Accuracy of Prognostic Classification
Next, we evaluated how the computer-assisted approach influenced determination of values below and above the cutoff of MC ≥ 5 for tumor prognostication (published cutoff for the MC as a solitary prognostic parameter). 8,35,42 For all pathologists and all 50 cases combined, 362, 170, and 116 instances were below and 788, 980, and 1034 instances were above the cutoff in stages 1, 2, and 3, respectively (Supplemental Table S5). Compared to the pHH3-assisted ground truth MCs (available for 40 cases), accuracy of classification below or above the cutoff, respectively, was 75.8% (95% CI: 72.3% to 80.1%) for stage 1, 86.6% (95% CI: 84.2% to 89.7%) for stage 2, 91.7% (95% CI: 89.9% to 94.0%) for stage 3, and 95% for the deep learning–based algorithm (algorithmic predictions without pathologist review). The increase of accuracy of the participants between stages 1 and 2 (P < .0001), stages 2 and 3 (P = .0012) and stages 1 and 3 (P < .0001) were significant (generalized linear mixed model). Due to the case selection strategy, the distribution of the hotspot MC values of the study cases is not representative of a routine diagnostic situation. We therefore divided the 40 cases into 5 tiers based on the pHH3-assisted ground truth MCs and determined individual accuracy values for these groups (Table 1). We determined that cases with ground truth MCs around the prognostic cutoff value had the lowest accuracy. However, also cases with pHH3-assisted ground truth MCs between 25 and 49 had falsely low MCs in 20/138 instances (14.5%) without computer assistance (stage 1), while 9/138 (6.5%) and 1/138 (0.7%) instances of those cases were misclassified in stages 2 and 3, respectively.
Table 1.
Accuracy of the 23 study participants (stages 1, 2, and 3) and the deep learning–based algorithm (without pathologist review) to classify mitotic counts (MC) as below (MC < 5) or above (MC ≥ 5) the prognostic cutoff as compared to the pHH3-assisted ground truth MC (GT-MC).
| GT-MC | Number of cases | Accuracy for | Stage 1a | Stage 2 | Stage 3 | Algorithm |
|---|---|---|---|---|---|---|
| 0–4 | 4 | Below cutoff | 75.0% | 50.0% | 50.0% | 50% |
| 5–9 | 7 | Above cutoff | 31.7% | 70.2% | 82.6% | 100% |
| 10–24 | 8 | Above cutoff | 63.0% | 89.1% | 99.5% | 100% |
| 25–49 | 6 | Above cutoff | 85.5% | 93.5% | 99.3% | 100% |
| ≥50 | 15 | Above cutoff | 99.4% | 100% | 100% | 100% |
| All cases | 40 | Below/above cutoff | 75.8% | 86.7% | 91.7% | 95% |
a The GT-MC and the participants’ MC of stage 1 were not determined in the same tumor location. The GT-MC and the MC of stages 2 and 3 were determined in the mitotic hotspot location based on the algorithmic heatmap.
Algorithmic Area Preselection Is Superior in Finding Mitotic Hot Spots
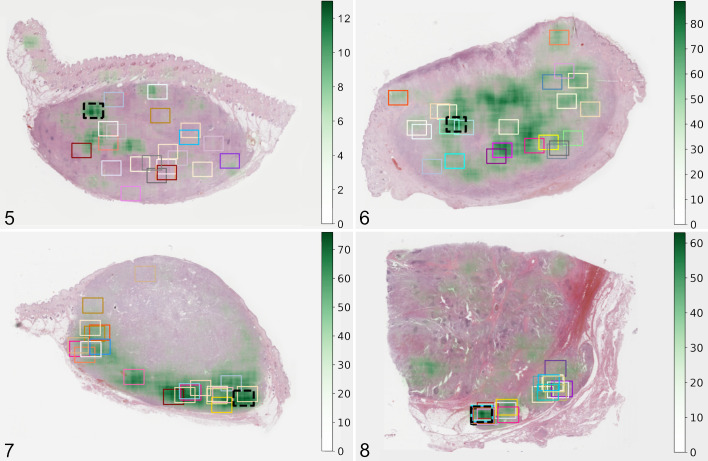
We evaluated the distribution of the manually selected MC-ROIs in stage 1 by the 23 participants. Visual assessment revealed that the approximate MC-ROIs were widely distributed throughout the tumor section in most cases, even if a high variability of the MC distribution (based on the algorithmic predictions) was present, that is, mitotic hotspots were not detected consistently (Figs. 5 –7, Suppl. Figs. S5–S54). In only 2 cases, a similar tumor area was consistently chosen for the MC-ROI. In one of these cases (No. 5; Fig. 8), a region within the tumor exhibiting local invasion was selected by all participants, and in the other case (No. 37; Suppl. Fig. S41), the tumor location with highest cellular density was selected by 22/23 participants. This contrasts with automated image analysis that will always propose the same tumor area (100% intra-algorithmic reproducibility).
Figures 5–8.
Approximate location of the mitotic count region of interest (MC-ROI) selected manually by each study participant (represented by the rectangular boxes) in the whole-slide images. The black box with the dashed line represents the algorithmically preselected MC-ROIs (algorithmic hotspot). The estimated MC heatmap is visualized by variable opacity of a green overlay (scale on the right side of image) on the histological image (hematoxylin and eosin stain) and is based on algorithmic mitotic figure predictions. Dark green areas represent mitotic hotspots. Figure 5. Case no. 33 with widely distributed MC-ROIs. Figure 6. Case no. 46 with widely distributed MC-ROIs. Figure 7. Case no. 38 with MC-ROIs mostly along the tumor periphery. Figure 8. Case no. 5 with similar MC-ROIs at a site of local tumor invasion.
As it was the goal to find mitotic hotspots and we hypothesized that computer assistance is helpful for this, we compared the MCs of the participants in the manually selected MC-ROIs (stage 1) and algorithmically preselected MC-ROIs (stage 2). Of the 1150 MC pairs from all participants, in 908 instances (79.0%), the MCs were higher in stage 2, and in 55 instances (4.8%), the MC was the same in stages 1 and 2. MCs of stage 1 were higher in 187/1150 MC pairs (16.2%) for all participants combined and in 0 to 20/50 MC pairs (median: 8; mean: 8.1) for each individual participant. For 151/187 of those instances with higher MCs in stage 1, it was possible to perform pHH3-assisted MCs for the MC-ROIs of both stages in order to more definitively determine which region had the higher mitotic density. The pHH3-assisted MC was slightly higher in the manually selected MC-ROI from stage 1 in one instance (0.7%), the same in both stages in 4 instances (2.6%) and higher in the algorithmically preselected MC-ROIs from stage 2 in 146 instances (96.7%). Only for one case (no. 27) was the algorithmically preselected MC-ROIs considered inappropriate as it composed of a tumor area with crush artifact that resulted in many false-positive predictions (pHH3-assisted MC was not available for this case).
Computer-Assisted MF Detection Improves Recall
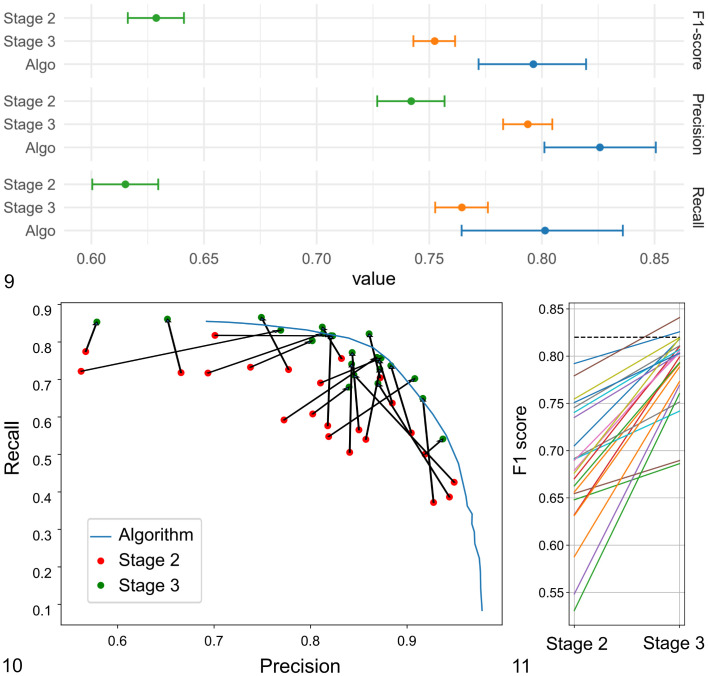
The ability of the participants to identify and classify individual MFs was determined for the same MC-ROIs in stage 2 (unaided MF identification) and stage 3 (computer-assisted MF identification). Annotations of each participant and predictions of the algorithm were compared to the pHH3-assisted ground truth. For the participants, we found an overall decrease of FNs by 38.4% between stage 2 (N = 23 107) and stage 3 (N = 14 256) and an overall increase of TPs by 23.7% between stage 2 (N = 37 117) and stage 3 (N = 45 929). A decrease of FNs and increase of TPs was present for 22/23 participants and a negligible decline (by 3 annotations; <1%) was present for the participant that had the lowest number of FN and highest number of TP in stage 2 (Supplemental Tables S6 and S7). FPs had an overall decrease by 3.8% between stage 2 (N = 10 981) and stage 3 (N = 10 582), whereas FPs were lower for 9 participants and higher for 14 participants in stage 3. Subsequently, the macro-averaged recall had an overall increase by 14.6 percentage points (maximum 30 percentage points; 22/23 participants improved), the macro-averaged precision had an overall increase of 2.3 percentage points (maximum 21 percentage points; 11/23 participants improved) and the macro-averaged F1-score had an overall increase of 10.7 percentage points (maximum of 22.9 percentage points; 23/23 participants improved) between stages 2 and 3 (Table 2, Figs. 9 –11). Differences between the 2 examination stages of the micro-averaged F1-score (12.4 percentage points), precision (5.2 percentage points), and recall (14.9 percentage points) were similar (Table 2). The 95% CIs for the micro-averaged F1-score, recall, and precision did not overlap between stages 2 and 3 (Fig. 9); thus, the improvement was considered significant. Experience level did not have an appreciable effect on participant performance (Supplemental Table S8). The deep learning–based algorithm (unverified predictions) had an almost balanced proportion of FP detections (N = 406) and FN detections (N = 496) and subsequently similar values for precision (0.84), recall (0.81), and the F1-score (0.83). The overall performance (F1-score) of the algorithms was comparatively high as it was not reached by participants in stage 2 and was only slightly exceeded by 2 participants in stage 3 (Fig. 11). The F1-score of the algorithm was considered to be significantly better than the score of the participants of both stages (95% CI did not overlap).
Table 2.
Performance (macro- and micro-averaged metrics with range or 95% confidence interval [CI]) of the 23 participants (partially or fully computer-assisted of stages 2 and 3, respectively) and the deep learning–based algorithm (unverified predictions) for detecting individual mitotic figures in mitotic hotspot regions of interest compared to a pHH3-assisted ground truth.
| Metrics | Precisiona | Recalla | F1 Scorea | |||
|---|---|---|---|---|---|---|
| Examination stage | 2 | 3 | 2 | 3 | 2 | 3 |
| Participants, macro-averaged values (range) | 0.80 (0.56–0.95) | 0.83 (0.58–0.94) | 0.62 (0.37–0.82) | 0.76 (0.54–0.87) | 0.68 (0.53–0.79) | 0.79 (0.67–0.84) |
| Participants, micro-averaged values (95% CI) | 0.74 (0.72–0.76) | 0.79 (0.78–0.81) | 0.62 (0.60–0.63) | 0.76 (0.75–0.78) | 0.63 (0.61–0.64) | 0.75 (0.74–0.77) |
| DL algorithm, macro-averaged value | 0.84 | 0.81 | 0.83 | |||
| DL algorithm, micro-averaged value (95% CI) | 0.83 (0.80–0.85) | 0.80 (0.76–0.84) | 0.80 (0.77–0.82) | |||
Abbreviations: DL, deep learning.
a The F1-score is the harmonic mean of precision (also known as positive predictive value) and recall (also known as sensitivity). The performance of the unverified algorithm is the same for stages 2 and 3.
Figures 9–11.
Object detection performance (identification and classification of mitotic figures) of the 23 participants and the deep learning–based algorithm in stages 2 and 3. Figure 9. Micro-averaged F1-score (upper graph), precision (middle graph), and recall (lower graph) with their 95% confidence intervals. The difference is considered significant if the intervals do not overlap. Figure 10. Macro-averaged recall and precision for the individual participants in stages 2 and 3 (connected by a black arrow) and the precision-recall curve for the algorithm (at different classification thresholds). For the algorithmic predictions for the present study, a single classification threshold was used that resulted in a recall of 0.81 and precision of 0.84. Figure 11. Macro-averaged F1-scores for the individual participants for stages 2 and 3. The dashed black line represents the F1-score of the algorithmic predictions.
Participants Considered Algorithmic MC-ROI Preselection to Be the Most Helpful Feature
Twenty-one of the 23 study participants (91%) filled out the concluding survey after they finished stage 3. Analysis revealed that participants considered MC-ROI selection to be the most difficult aspect of performing the manual MC (stage 1), whereas spotting potential MF candidates and classifying them against lookalikes was considered comparably easy (Supplemental Tables S9 and S10 and Fig. S55). Subsequently, most participants had the subjective impression that algorithmic MC-ROI preselection was extremely or very helpful. In contrast, visualization of algorithmic predictions, as well as display of their algorithmic confidence values, was generally considered helpful to a lesser degree (Supplemental Tables S11 and S12 and Fig. S56). Almost all participants (N = 20/21) indicated that their decision of classifying MFs against lookalikes was consciously influenced in stage 3 by the algorithmic confidence value at variable degrees ranging from being influenced in very few to many potential MF candidates (Supplemental Table S13), especially if participants were uncertain about the MF candidate (N = 17/20). Most participants considered digital microscopy slightly (N = 12/21) or significantly (N = 3/15) inferior to light microscopy for identification of MF (Supplemental Table S14) and mostly deemed fine-focusing/z-stacking (not available for this study) necessary for at least some MF candidates (N = 14/21; Supplemental Table S15). The majority of the participants (81%; N = 17/21) considered the rectangular shape of the MC-ROI to be acceptable for performing the MC in the study cases, while 2 participants found the shape inappropriate (2 participants had no opinion).
Discussion
The present study confirmed that inconsistency and inaccuracy of the MC arises from a combination of inappropriate MC-ROI selection (failure to find mitotic hot spots), incomplete MF identification, and imprecise MF classification. In order to enable highest prognostic value of the MC, it is necessary to develop methods to improve those critical steps. We addressed each of the 3 critical steps of the MC with our computer-assisted approaches via algorithmic area preselection (stage 2), MF candidate visualization (stage 3), and display of algorithmic confidence values (stage 3). We were able to demonstrate significantly increased performance on the overall MC level and individual MF level with computer assistance. Participants of the present study reported that screening tumor sections for mitotic hotspots is the most difficult task of manual MCs and subsequently deemed algorithmic MC-ROI preselection by far the most useful tool. In fact, we have shown that algorithmic area preselection was superior to manual area selection in almost all instances. Nevertheless, visualization of potential MF candidates also had a strong positive effect on the pathologists’ ability to find MFs in MC-ROIs (recall). In contrast, the benefit of displaying the algorithmic confidence values was considered controversial by participants (see below) and did not seem to have a strong positive impact on precision.
The results of the present study reveal high variability of the MC between pathologists that has led to marked inconsistency in prognostic stratification (in our case MC < 5 vs MC ≥ 5 8,35,42 ). While it was beyond the scope of the present study to correlate the different MC methods with patient outcome, it is assumed that this degree of inconsistency might have a relevant influence on the appropriate treatment recommendation by clinicians or oncologists. Of note, computer assistance has led to a more consistent classification of the cases into the prognostic cutoff ranges, but we have also found an overall higher MC for our computer-assisted methods. Therefore, new prognostic cutoff values and stratifications have to be determined, potentially for each individual computer-assisted approach and software tool. For example, Elston et al 19 have proposed a cutoff of 0 for group 1 ccMCTs with good prognosis (group 2: 1–7; group 3: >7). This prognostic group is notably reduced in larger tumor sections (by a factor of 21×) if full computer assistance (stage 3) is used due to the improved sensitivity of MF detection. A limitation of the present study is that only a few cases with a truly low MC (below cutoff value) were included, and future studies need to verify our results for this subgroup. Future studies are also needed to evaluate the prognostic capability of computer-assisted MCs.
A particular strength of the present study was the use of a pHH3-assisted ground truth for performance evaluation. Most previous studies have evaluated their MF algorithms against a majority vote of pathologists’ annotations from HE images, 3,5,12,33,38,45,47 which might be problematic due to human limitations in performing this task. If a consensus is generated by such a large group of participants as in the present study, a majority vote is very likely to include predominantly clear MFs and would not contain many morphologically inconclusive or equivocal but still “true” MFs. Hence, a majority vote will not necessarily represent the biological truth. 10 The advantage of pHH3 immunohistochemistry (as opposed to HE images) is that MFs are easily spotted (higher sensitivity) and more easily classified against lookalikes (higher specificity with the exception of telophase MF). 17,20,36 We highlight that some of the annotated pHH3-positive cells were extremely difficult to classify as MFs in the HE image (especially early prophase or if cells were tangentially sectioned) and might therefore have been missed (false negative) by study pathologists in all MC approaches. It is acknowledged that pHH3-derived labeling may overestimate prophase MFs and underestimate telophase MFs as compared to solely HE-based labeling. 39 Therefore, we decided to use a combination of the pHH3 and HE image for annotating the ground truth. We believe that our ground truth labeling approach is the most objective and accurate reference (gold standard) method available for MFs to date and was independent of the participants (as opposed to a majority vote ground truth). The ground truth dataset was annotated by the principal investigator, who was not involved as a study participant and was only partially responsible for creating the training dataset of the deep learning model (labels were created by consensus of 2 pathologists). Also, pHH3-immunolabeling was not available for the study participants nor for development of the algorithm. Regardless, the pHH3-assisted ground truth is still observer-dependent and therefore not errorless, and a bias on performance evaluation cannot be complete ruled out.
We have shown that identification of mitotic hotspots can be tremendously improved with computer assistance. 5 An advantage of algorithms is that they can efficiently analyze entire WSIs with 100% intra-algorithmic reproducibility and are therefore able to determine the mitotic distribution consistently in entire or even multiple tumor sections, while manual MF screening by pathologists is restricted to some field of views at high magnification due to time constraints. A limitation of our deep learning method was that 10% of the cases analyzed had an algorithmic MC-ROI preselection outside of the tumor area (cases were excluded) and one case had an algorithmic MC-ROI preselection in an area of crush artifacts. A previous study reported inappropriate MC-ROI selection in 58% of the cases. 7 In the present study, participants were unable to change the preselected MC-ROI location, which was necessary in order to determine and compare the pathologist’s performance for detecting individual MFs. However, we highlight that software for routine application should allow the pathologist to modify the MC-ROI location if the preselected MC-ROI is considered inappropriate due to algorithmic failure. While it was beyond the scope of the present study to investigate approaches for software-pathologist interaction on MC-ROI selection, we propose that computer-assisted MC tools include one of the following features: (1) restriction of the algorithmic predictions to the actual tumor area, which can be delineated manually by a pathologist 7 or segmented automatically by means of deep learning–based algorithms; 22 (2) heat map visualization of mitotic density for quick manual selection and correction of the MC-ROI; 15 (3) proposal of the top 3 to 5 hotspots areas from which pathologists can choose. Another aspect that needs to be considered for future implementation of computer-assisted approaches is the shape of the MC-ROI. Even though most participants found the rectangular shape appropriate for the examined ccMCT cases, it might be useful to be able to adjust the MC-ROI shape for tumor types that require exclusion of relevant non-neoplastic tissue (necrosis, hair follicles, connective tissue, etc) from the MC-ROI. 18,32
Even if pathologists examine the same image sections, variability of the number of enumerated MFs has been noted, 16,44,45,47 and computer assistance seems to be a promising solution for improving MF identification and classification. Similar to the comparison between stages 2 and 3 of the present study, Pantanowitz et al 33 investigated the influence of computer assistance on the pathologist’s performance of annotating MFs in “hpf” images. Pathologists in that study 33 had a somewhat lower performance (F1-score) overall as compared to our results (7.1% lower without and 7.7% lower with computer assistance), which might be related to the ground truth definition used. Pantanowitz et al 33 used the majority vote of 4/7 pathologists to define a “true” MF (see above). Nevertheless, Pantanowitz et al 33 demonstrated an increase of the overall F1-score similar to that of the present study (10% vs 10.7%); however, they had a slightly lower increase of the overall recall (11.7% vs 14.6%) and a higher increase of overall precision (8.8% vs 2.3%) than our study. This might also be related to the ground truth definition used, or the performance of the image analysis algorithm applied, or due to the level of the individual pathologists’ acceptance of the algorithmic predictions.
Of note, our results demonstrate that individual pathologists may have high recall/low precision, moderate recall/moderate precision, or high precision/low recall. This is probably influenced to a large part by the decision criteria of individual pathologists for ambiguous patterns, and standardized morphological criteria for MFs might be helpful to harmonize the pathologist’s decision. 18 Although all pathologists had overall high performance (F1-score), the individual precision-recall tradeoff of different pathologists may have a tremendous influence on the MCs and the prognostic stratification based on specific cutoff values. Our results show that the computer-assisted approach (stage 3) generally shifted the individual pathologists somewhat toward a more harmonic tradeoff between recall and precision (moderate recall/moderate precision). The overall direction of the shift coincides with the precision/recall of the algorithmic results and could be influenced by a confirmation bias of the experts. Unlike the study by Pantanowitz et al, 33 we supplied the study participants with the algorithmic confidence value in stage 3. Although the model confidence scores overall had a strong correlation with the ground truth, some participants commented that a few confidence values contrasted with their degree of certainty. Interestingly, many participants felt that they were (negatively) biased by the confidence values especially for difficult MF candidates. A previous study has in fact shown that pathologist may fail to identify a high proportion of incorrect (false negative and false positive) algorithmic predictions, 29 and future studies need to determine the (positive or negative) effect of this bias in a diagnostic setting.
The results of our study were highly dependent upon adequate performance of the applied algorithm. Algorithms always exhibit 100% intra-algorithmic reproducibility for the same images, but accuracy may vary largely between different algorithms (inter-algorithmic variability; depending on numerous factors). The algorithm used in this study was created and evaluated on a distinct dataset without employing IHC labeling. Yet, the algorithmic predictions of the present study were highly accurate (F1-score) compared to the pHH3-assisted ground truth (as compared to the study participants). The high performance of the algorithm used for the present study can be attributed to the advanced deep learning methods applied, 5 the high quality and quantity of the dataset used for training of the algorithm, 12 and the high representativeness of the training dataset for the present study cases (little domain shift, see below). However, we also experienced several incorrect MF predictions and some inappropriate algorithmic MC-ROI preselections, which necessitates review by trained experts (computer-assisted MC) and further research on algorithmic solutions.
One of the most relevant concerns that cannot be considered solved to date is robustness in the application of such algorithms on images that are acquired with other scanners or have strong differences in HE staining (domain shift). 3,4,10 The consequence is that algorithms cannot necessarily be applied to images from different laboratories without prior validation and possibly modification (such as by color normalization and augmentation, 39,44 domain adaptation, 4 and/or transfer learning 3 ). A limitation of deep learning–based models is that the specific decision criteria are mostly not transparent (“black box”) and situations in which the algorithm may fail are not easily foreseeable. It is those “unexpected” situations that highlight the necessity of careful validation of each new application for its intended use. Deep learning models can be tested statistically (using metrics such as recall, precision or the F1 score), 10 and each laboratory that wants to use a MF algorithm should validate its performance based on those metrics. The 50 cases included in the present study support the impression that the algorithm works reliably in many cases, yet the included cases may not represent all potential sources of error and had a similar domain as the images used for training the deep learning model. A recent pilot study has shown that expert review, that is, computer assistance as opposed to unverified algorithmic predictions, is necessary to ensure high reliability of the MC in cases that have a presumed marked domain shift (and thus higher rate of algorithmic errors). 15 Therefore, we consider computer-assisted approaches with verification by a trained pathologist to be highly beneficial, at least until more experience and progress is gained through research and routine use of these applications. Access to intermediate results of the algorithmic pipeline (such as visualization of MF predictions) may allow a higher degree of comprehensibility by the reviewing pathologist and should therefore be encouraged. Future studies need to determine which degree of expert review is required, how reliably pathologists can detect algorithmic errors, and to what degree the decision of pathologists is consciously and unconsciously biased by algorithmic predictions.
If used reasonably, computer-assisted MC may also have a positive influence on the time required for examination of tumor specimens. For example, visualization of algorithmic MF detections in WSI has been reported to significantly reduce time required for MF enumeration, which was especially notable for less experienced pathologists. 29,33 A recent pilot study has also reported a significant time improvement when using MC heat maps and MF candidate visualizations as a guide for a pathologist. 15 In the present study, participants noted an enormous time improvement between stages 1 and 2 due to omission of manual MC-ROI selection. However, we decided not to measure labor intensity of the 3 stages systematically, as our focus was to evaluate the potential benefits, limitations, and requirements of these software tools. Future studies can scrutinize the most efficient and reliable workflow when using these tools in a realistic diagnostic situation.
A limitation of the present study was that the algorithmically preselected MC-ROI could not be corrected and each MF had to be manually annotated in order to be able to calculate the performance of MF detection in the same tumor areas. This limitation does not reflect a realistic diagnostic situation that, on the one hand, might require manual correction of inappropriately preselected MC-ROIs (see above). On the other hand, time investment could possibly be reduced if pathologists only had to add MF missed by the algorithm and remove wrong predictions instead of annotating all MF (including the correct algorithmic predictions), as in the present study. Efficiency of computer-assisted diagnosis/prognosis largely depends on the degree of computer assistance (eg, stage 2 vs stage 3), the extent of pathologist’s review of algorithmic predictions (verify a subset or all predictions or even only a specific subset of cases such as borderline cases), the frequency of algorithmic errors (and thus the required extent of manual corrections), ease of use of the developed software (pathologist-software interaction), the pathologist’s experience with the computer-assisted diagnosis/prognosis and the specific software, and the way the software is incorporated into the diagnostic workflow. Future studies should focus on these aforementioned aspects and thereby determine an appropriate tradeoff between workflow efficiency and diagnostic reliability for a diagnostic setting. Future software development should improve on the assistive visualization of algorithmic predictions. For example, an assembly with small image tiles of all individual MF predictions from the (pre)selected MC-ROI in a separate viewing window could possibly allow a quick review of predictions (true positives vs false positives) without having to navigate through the MC-ROI as has been described for automated blood smear evaluation systems. 40
Conclusion
Our results demonstrate that computer assistance using an accurate deep learning–based model is a promising method for improving reproducibility and accuracy of MCs in histological tumor sections. Full computer assistance (assistance in MC-ROI selection and MF identification and classification) was superior to partial computer assistance (only assistance in MC-ROI selection) in the present study. This study shows that computer-assisted MCs may be a valuable method for standardization in future research studies and routine diagnostic tumor assessment using digital microscopy. Furthermore, improved work efficiency (such as by MC-ROI preselection) may be of interest for diagnostic laboratories and additional studies need to evaluate the degree of verification of algorithmic predictions required. Future research should also evaluate whether computer-assisted MC approaches will benefit tumor prognostication (compared to patient outcome).
Supplemental Material
Supplemental Material, sj-pdf-1-vet-10.1177_03009858211067478 for Computer-assisted mitotic count using a deep learning–based algorithm improves interobserver reproducibility and accuracy by Christof A. Bertram, Marc Aubreville, Taryn A. Donovan, Alexander Bartel, Frauke Wilm, Christian Marzahl, Charles-Antoine Assenmacher, Kathrin Becker, Mark Bennett, Sarah Corner, Brieuc Cossic, Daniela Denk, Martina Dettwiler, Beatriz Garcia Gonzalez, Corinne Gurtner, Ann-Kathrin Haverkamp, Annabelle Heier, Annika Lehmbecker, Sophie Merz, Erica L. Noland, Stephanie Plog, Anja Schmidt, Franziska Sebastian, Dodd G. Sledge, Rebecca C. Smedley, Marco Tecilla, Tuddow Thaiwong, Andrea Fuchs-Baumgartinger, Donald J. Meuten, Katharina Breininger, Matti Kiupel, Andreas Maier and Robert Klopfleisch in Veterinary Pathology
Acknowledgement
We thank Nicole Huth for technical support.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Christof A. Bertram gratefully acknowledges financial support received from the Dres. Jutta und Georg Bruns-Stiftung für innovative Veterinärmedizin.
ORCID iDs: Christof A. Bertram  https://orcid.org/0000-0002-2402-9997
https://orcid.org/0000-0002-2402-9997
Marc Aubreville  https://orcid.org/0000-0002-5294-5247
https://orcid.org/0000-0002-5294-5247
Taryn A. Donovan  https://orcid.org/0000-0001-5740-9550
https://orcid.org/0000-0001-5740-9550
Alexander Bartel  https://orcid.org/0000-0002-1280-6138
https://orcid.org/0000-0002-1280-6138
Kathrin Becker  https://orcid.org/0000-0002-0922-8189
https://orcid.org/0000-0002-0922-8189
Martina Dettwiler  https://orcid.org/0000-0001-9404-7122
https://orcid.org/0000-0001-9404-7122
Annabelle Heier  https://orcid.org/0000-0003-0915-4236
https://orcid.org/0000-0003-0915-4236
Rebecca C. Smedley  https://orcid.org/0000-0001-5704-2664
https://orcid.org/0000-0001-5704-2664
Robert Klopfleisch  https://orcid.org/0000-0002-6308-0568
https://orcid.org/0000-0002-6308-0568
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Al-Janabi S, van Slooten HJ, Visser M, et al. Evaluation of mitotic activity index in breast cancer using whole slide digital images. PLoS One. 2013;8(12):e82576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aubreville M, Bertram C, Klopfleisch R, et al. SlideRunner—a tool for massive cell annotations in whole slide images. In: Maier, Deserno A, Handels TMH, eds. Bildverarbeitung Für Die Medizin 2018. Springer; 2018:309–314. [Google Scholar]
- 3. Aubreville M, Bertram CA, Donovan TA, et al. A completely annotated whole slide image dataset of canine breast cancer to aid human breast cancer research. Sci Data. 2020;7(1):417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aubreville M, Bertram CA, Jabari S, et al. Inter-species, inter-tissue domain adaptation for mitotic figure assessment—learning new tricks from old dogs. In: Tolxdorff, Deserno T, Handels TMH, eds. Bildverarbeitung Für Die Medizin 2020: Springer Vieweg; 2020:1–7. [Google Scholar]
- 5. Aubreville M, Bertram CA, Marzahl C, et al. Deep learning algorithms out-perform veterinary pathologists in detecting the mitotically most active tumor region. Sci Rep. 2020;10(1):16447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Avallone G, Rasotto R, Chambers JK, et al. Review of histological grading systems in veterinary medicine. Vet Pathol. 2021;58(5):809–828. [DOI] [PubMed] [Google Scholar]
- 7. Balkenhol MCA, Tellez D, Vreuls W, et al. Deep learning assisted mitotic counting for breast cancer. Lab Invest. 2019;99(11):1596–1606. [DOI] [PubMed] [Google Scholar]
- 8. Berlato D, Murphy S, Laberke S, et al. Comparison of minichromosome maintenance protein 7, Ki67 and mitotic index in the prognosis of intermediate Patnaik grade cutaneous mast cell tumours in dogs. Vet Comp Oncol. 2018;16(4):535–543. [DOI] [PubMed] [Google Scholar]
- 9. Berlato D, Murphy S, Monti P, et al. Comparison of mitotic index and Ki67 index in the prognostication of canine cutaneous mast cell tumours. Vet Comp Oncol. 2015;13(2):143–150. [DOI] [PubMed] [Google Scholar]
- 10. Bertram CA, Aubreville M, Donovan TA, et al. International guidelines for veterinary tumor pathology: a call to action; Guideline 11.0: Computational pathology for tumor histopathology. Vet Pathol. 2021;58(5):Supplemental material. [DOI] [PubMed] [Google Scholar]
- 11. Bertram CA, Aubreville M, Gurtner C, et al. Computerized calculation of mitotic count distribution in canine cutaneous mast cell tumor sections: mitotic count is area dependent. Vet Pathol. 2020;57(2):214–226. [DOI] [PubMed] [Google Scholar]
- 12. Bertram CA, Aubreville M, Marzahl C, et al. A large-scale dataset for mitotic figure assessment on whole slide images of canine cutaneous mast cell tumor. Sci Data. 2019;6(1):274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bertram CA, Gurtner C, Dettwiler M, et al. Validation of digital microscopy compared with light microscopy for the diagnosis of canine cutaneous tumors. Vet Pathol. 2018;55(4):490–500. [DOI] [PubMed] [Google Scholar]
- 14. Bertram CA, Klopfleisch R. The pathologist 2.0: an update on digital pathology in veterinary medicine. Vet Pathol. 2017;54(4):756–766. [DOI] [PubMed] [Google Scholar]
- 15. Bertram CA, Klopfleisch R, Bartel A, et al. Expert review of algorithmic mitotic count predictions ensures high reliability. In: 4th ESVP, ECVP and ESTP Cutting Edge Pathology Congress. Virtual Congress; September 15-17, 2021. [Google Scholar]
- 16. Bertram CA, Veta M, Marzahl C, et al. Are pathologist-defined labels reproducible? Comparison of the TUPAC16 mitotic figure dataset with an alternative set of labels. In: Cardoso J, ed. Interpretable and Annotation-Efficient Learning for Medical Image Computing. iMIMIC 2020/MIL3iD 2020/LABELS 2020. Vol 12446. Springer Nature; 2020:204–213. [Google Scholar]
- 17. Cui X, Harada S, Shen D, et al. The utility of phosphohistone H3 in breast cancer grading. Appl Immunohistochem Mol Morphol. 2015;23(10):689–695. [DOI] [PubMed] [Google Scholar]
- 18. Donovan TA, Moore FM, Bertram CA, et al. Mitotic figures-normal, atypical, and imposters: a guide to identification. Vet Pathol. 2021;58(2):243–257. [DOI] [PubMed] [Google Scholar]
- 19. Elston LB, Sueiro FA, Cavalcanti JN, et al. The importance of the mitotic index as a prognostic factor for survival of canine cutaneous mast cell tumors: a validation study. Vet Pathol. 2009;46(2):362–365. [DOI] [PubMed] [Google Scholar]
- 20. Focke CM, Finsterbusch K, Decker T, et al. Performance of 4 immunohistochemical phosphohistone H3 antibodies for marking mitotic figures in breast cancer. Appl Immunohistochem Mol Morphol. 2018;26(1):20–26. [DOI] [PubMed] [Google Scholar]
- 21. Greenland S, Senn SJ, Rothman KJ, et al. Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur J Epidemiol. 2016;31(4):337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo Z, Liu H, Ni H, et al. A fast and refined cancer regions segmentation framework in whole-slide breast pathological images. Sci Rep. 2019;9(1):882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hendzel MJ, Wei Y, Mancini MA, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106(6):348–360. [DOI] [PubMed] [Google Scholar]
- 24. Horta RS, Lavalle GE, Monteiro LN, et al. Assessment of canine mast cell tumor mortality risk based on clinical, histologic, immunohistochemical, and molecular features. Vet Pathol. 2018;55(2):212–223. [DOI] [PubMed] [Google Scholar]
- 25. Jannink I, van Diest PJ, Baak JP. Comparison of the prognostic value of four methods to assess mitotic activity in 186 invasive breast cancer patients: classical and random mitotic activity assessments with correction for volume percentage of epithelium. Hum Pathol. 1995;26(10):1086–1092. [DOI] [PubMed] [Google Scholar]
- 26. Jiang J, Larson NB, Prodduturi N, et al. Robust hierarchical density estimation and regression for re-stained histological whole slide image co-registration. PLoS One. 2019;14(7):e0220074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kiupel M, Webster JD, Bailey KL, et al. Proposal of a 2-tier histologic grading system for canine cutaneous mast cell tumors to more accurately predict biological behavior. Vet Pathol. 2011;48(1):147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marzahl C, Aubreville M, Bertram CA, et al. Deep learning-based quantification of pulmonary hemosiderophages in cytology slides. Sci Rep. 2020;10(1):9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marzahl C, Bertram CA, Aubreville M, et al. Are fast labeling methods reliable? A case study of computer-aided expert annotations on microscopy slides. In: Martel, Abolmaesumi AL, Stoyanov PD, eds. Medical Image Computing and Computer Assisted Intervention—MICCAI 2020. Springer; 2020:24–32. [Google Scholar]
- 30. McAlpine ED, Michelow P. The cytopathologist’s role in developing and evaluating artificial intelligence in cytopathology practice. Cytopathology. 2020;31(5):385–392. [DOI] [PubMed] [Google Scholar]
- 31. Meuten D, Moore F, Donovan T, et al. International guidelines for veterinary tumor pathology: a call to action. Vet Pathol. 2021;58(5):766–794. [DOI] [PubMed] [Google Scholar]
- 32. Meuten D, Moore F, George J. Mitotic count and the field of view area: time to standardize. Vet Pathol. 2016;53(1):7–9. [DOI] [PubMed] [Google Scholar]
- 33. Pantanowitz L, Hartman D, Qi Y, et al. Accuracy and efficiency of an artificial intelligence tool when counting breast mitoses. Diagn Pathol. 2020;15(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Puri M, Hoover SB, Hewitt SM, et al. Automated computational detection, quantitation, and mapping of mitosis in whole-slide images for clinically actionable surgical pathology decision support. J Pathol Inform. 2019;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Romansik EM, Reilly CM, Kass PH, et al. Mitotic index is predictive for survival for canine cutaneous mast cell tumors. Vet Pathol. 2007;44(3):335–341. [DOI] [PubMed] [Google Scholar]
- 36. Skaland I, Janssen EA, Gudlaugsson E, et al. Phosphohistone H3 expression has much stronger prognostic value than classical prognosticators in invasive lymph node-negative breast cancer patients less than 55 years of age. Mod Pathol. 2007;20(12):1307–1315. [DOI] [PubMed] [Google Scholar]
- 37. Sledge DG, Webster J, Kiupel M. Canine cutaneous mast cell tumors: a combined clinical and pathologic approach to diagnosis, prognosis, and treatment selection. Vet J. 2016;215:43–54. [DOI] [PubMed] [Google Scholar]
- 38. Tabata K, Uraoka N, Benhamida J, et al. Validation of mitotic cell quantification via microscopy and multiple whole-slide scanners. Diagn Pathol. 2019;14(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tellez D, Balkenhol M, Otte-Höller I, et al. Whole-slide mitosis detection in H&E breast histology using PHH3 as a reference to train distilled stain-invariant convolutional networks. IEEE Trans Med Imaging. 2018;37:2126–2136. [DOI] [PubMed] [Google Scholar]
- 40. Tvedten HW, Lilliehöök IE. Canine differential leukocyte counting with the CellaVision DM96Vision, Sysmex XT-2000iV, and Advia 2120 hematology analyzers and a manual method. Vet Clin Pathol. 2011;40(3):324–339. [DOI] [PubMed] [Google Scholar]
- 41. van Lelyveld S, Warland J, Miller R, et al. Comparison between Ki-67 index and mitotic index for predicting outcome in canine mast cell tumours. J Small Anim Pract. 2015;56(5):312–319. [DOI] [PubMed] [Google Scholar]
- 42. Vascellari M, Giantin M, Capello K, et al. Expression of Ki67, BCL-2, and COX-2 in canine cutaneous mast cell tumors: association with grading and prognosis. Vet Pathol. 2013;50(1):110–121. [DOI] [PubMed] [Google Scholar]
- 43. Veta M, Heng YJ, Stathonikos N, et al. Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge. Med Image Anal. 2019;54:111–121. [DOI] [PubMed] [Google Scholar]
- 44. Veta M, van Diest PJ, Jiwa M, et al. Mitosis counting in breast cancer: object-level interobserver agreement and comparison to an automatic method. PLoS One. 2016;11(8):e0161286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Veta M, van Diest PJ, Willems SM, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20(1):237–248. [DOI] [PubMed] [Google Scholar]
- 46. Wei BR, Halsey CH, Hoover SB, et al. Agreement in histological assessment of mitotic activity between microscopy and digital whole slide images informs conversion for clinical diagnosis. Acad Pathol. 2019;6:2374289519859841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilm F, Bertram CA, Marzahl C, et al. Influence of inter-annotator variability on automatic mitotic figure assessment. In: Palm, Deserno C, Handels TMH, eds. Bildverarbeitung für die Medizin 2021. Springer Fachmedien Wiesbaden; 2021:241–246. [Google Scholar]
- 48. Zhang D, Wang J, Zhao X. Estimating the uncertainty of average F1 scores. In: Allan, Croft JB, eds. Proceedings of the 2015 International Conference on the Theory of Information Retrieval; Northampton, MA; September 27, 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-vet-10.1177_03009858211067478 for Computer-assisted mitotic count using a deep learning–based algorithm improves interobserver reproducibility and accuracy by Christof A. Bertram, Marc Aubreville, Taryn A. Donovan, Alexander Bartel, Frauke Wilm, Christian Marzahl, Charles-Antoine Assenmacher, Kathrin Becker, Mark Bennett, Sarah Corner, Brieuc Cossic, Daniela Denk, Martina Dettwiler, Beatriz Garcia Gonzalez, Corinne Gurtner, Ann-Kathrin Haverkamp, Annabelle Heier, Annika Lehmbecker, Sophie Merz, Erica L. Noland, Stephanie Plog, Anja Schmidt, Franziska Sebastian, Dodd G. Sledge, Rebecca C. Smedley, Marco Tecilla, Tuddow Thaiwong, Andrea Fuchs-Baumgartinger, Donald J. Meuten, Katharina Breininger, Matti Kiupel, Andreas Maier and Robert Klopfleisch in Veterinary Pathology