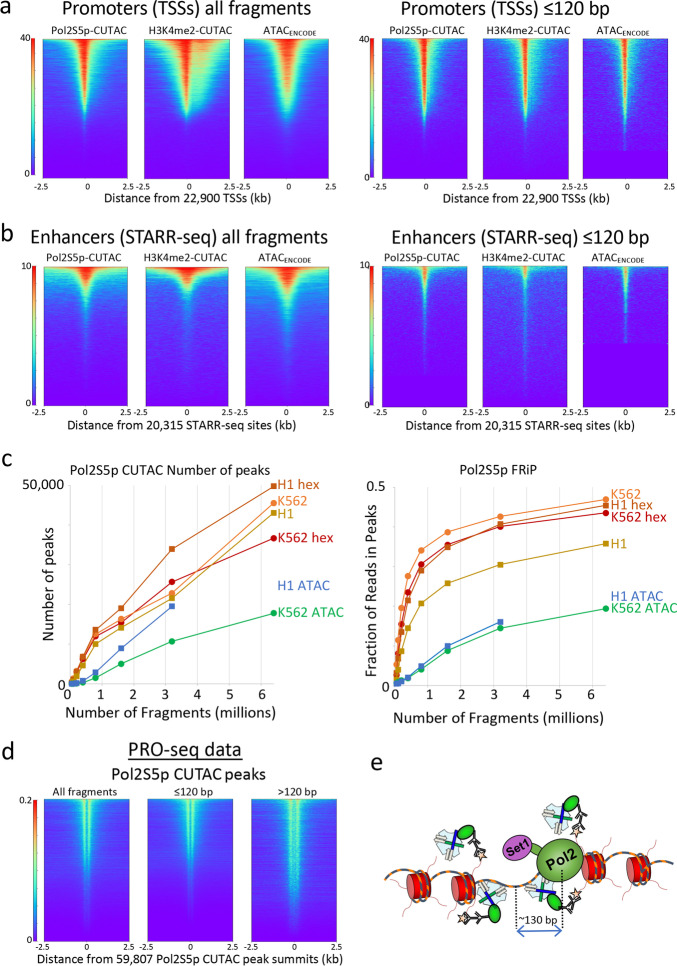
Fig. 1.
Pol2S5p CUTAC maps promoters and functional enhancers adjacent to RNAP2 genome-wide. a Heatmaps showing occupancies of Pol2S5p CUTAC, H3K4me2 CUTAC, and ATAC-seq signals over promoters in K562 cells, which become sharper for subnucleosomal (1–120 bp) fragments. b Pol2S5p CUTAC, H3K4me2 CUTAC, and ATAC-seq signals precisely mark functional enhancers when aligned to STARR-seq peaks. c To evaluate the data quality of Pol2S5p CUTAC, random samples of mapped fragments were drawn, mitochondrial reads were removed and MACS2 was used to call (narrow) peaks. The number of peaks called for each sample (left) is a measure of sensitivity, and the fraction of reads in peaks (FRiP, right) is a measure of specificity calculated for each sampling in a doubling series from 50,000 to 6.4 million fragments. For comparison, an ENCODE ATAC-seq sample was used for K562 cells and a published ATAC-seq sample from our lab (GSE128499) was used for H1 cells. Hex samples were tagmented in the presence of 10% 1,6-hexanediol. d Run-on transcription initiates from most sites corresponding to RNAP2S5p CUTAC peaks, where PRO-seq maps the RNA base at the active site of paused Pol2 [14]. Both plus and minus strand PRO-seq datasets downloaded from GEO (GSM3452725) were pooled and aligned over peaks called by MACS2 using 3.2 million RNAP2S5p CUTAC fragments. e Model for RNAP2S5p-tethered tagmentation of adjacent accessible DNA, where the Set1 H3K4 methyltransferase di- and tri-methylates nucleosomes near stalled Pol2