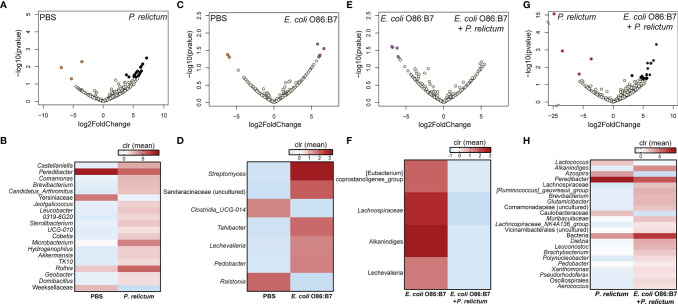
Figure 3.
Impact of E. coli O86:B7 vaccine and P. relictum infection on the taxonomic profiles of mosquito microbiota. Volcano plot showing the differential bacterial abundance in mosquito microbiota between different experimental groups (A) PBS vs. P. relictum, (C) PBS vs. E. coli O86:B7, (E) E. coli O86:B7 vs. E. coli O86:B7+P. relictum and (G) P. relictum vs. E. coli O86:B7+P. relictum. Taxa with significant differences (Wald test, p < 0.05) between the groups are represented with colored dots (i.e., orange, purple, red and black). The gray dots represent taxa with no significant differences between groups. Heatmaps representing the relative abundance (expressed as clr) of taxa with significant differences between (B) PBS vs. P. relictum, (D) PBS vs. E. coli O86:B7, (F) E. coli O86:B7 vs. E. coli O86:B7+P. relictum and (H) P. relictum vs. E. coli O86:B7+P. relictum. Taxa with significant differences in their abundance were identified using DeSeq2 algorithm. The taxonomic profiles were obtained from 16S rRNA sequences from midgut of mosquitoes fed on mock-immunized (n = 5 mosquitoes midgut pool), E. coli O86:B7-immunized (n = 5 mosquitoes midgut pool), E. coli O86:B7-immunized and challenged with P. relictum (n = 8 mosquitoes midgut pool) and challenged only with P. relictum (n = 8 mosquitoes midgut pool).