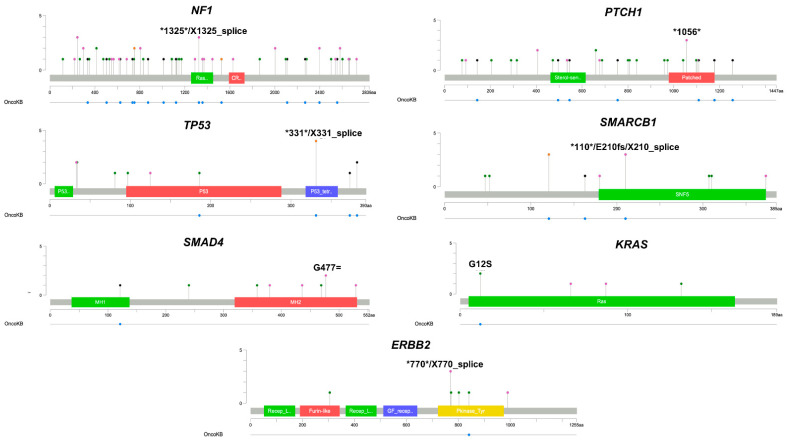
Figure 3.
Gene locations of somatic variants in oncogenes and tumor-suppressor genes with predicted oncogenic impact, as per OncoKB database. Seven oncogenes/tumor suppressor genes were identified with shared somatic variants detected in ≥2 CTCs. Lollipop plots show the variant gene locations and their predicted oncogenic impact, according to OncoKB database. In some genes, at the same base position different mutations are observed in multiple individual CTCs. For example, in NF1 we observed a base substitution in coding sequence position of 1325 that results in the introduction of a stop codon and, separately at the same base position, a base substitution occurs that alters the conserved splice acceptor/donor site for exon/intron splicing. Colored boxes represent mutations in specific functional domains (*: stop codon; mutation types: green: missense, black: truncating, orange: splice, pink: others).