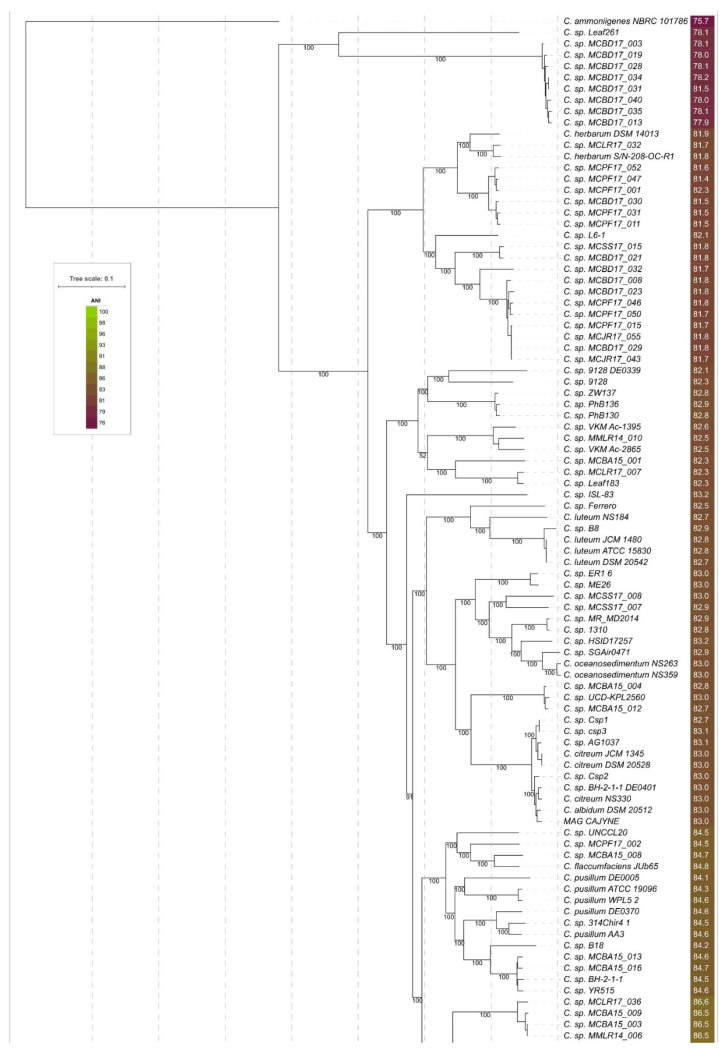
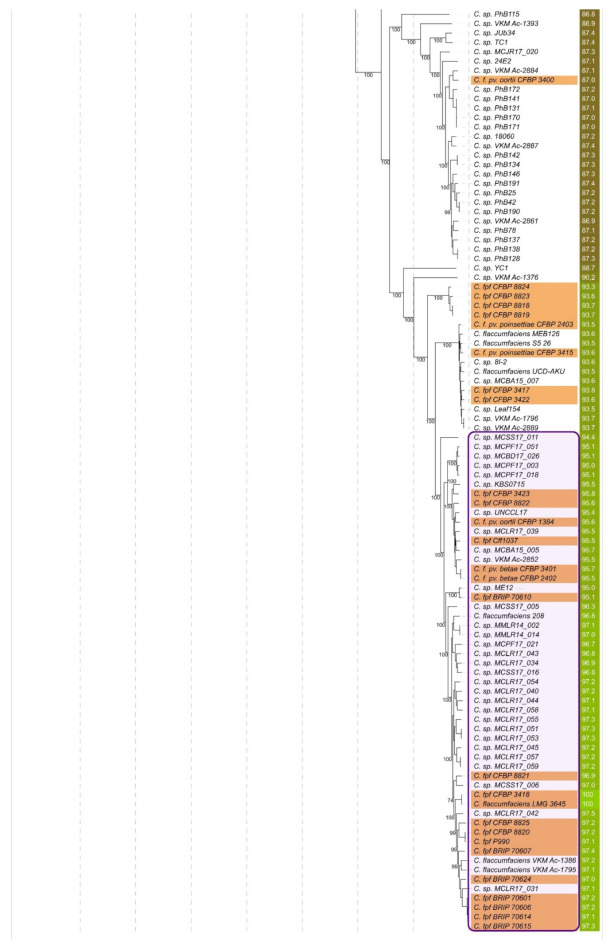
Figure 4.
Best-scoring phylogenetic trees obtained with RAxML using 190 Curtobacterium genomes. The abbreviations are as follows: C.—Curtobacterium, C. f.—Curtobacterium flaccumfaciens, and C. fpf—flaccumfaciens pv. flaccumfaciens. The C. fpf strains with confirmed pathogenicity are coloured yellow-orange. The group of 53 strains outlined with violet constitutes a possible reclassified species of C. flaccumfaciens, based on the ANI and phylogeny results. ANI values compared to the C. fpf CFBP 3418-type strain are shown to the right of the organism’s name and coloured according to a heat map scale. Bootstrap support values are shown near the branches of the rectangular tree as a percentage of 1000 replicates. The scale bar shows 0.1 estimated substitutions per site, and the tree was rooted to Curtobacterium ammoniigenes NBRC 101786.