Abstract
OBJECTIVE
Maturity-onset diabetes of the young (MODY) is frequently misdiagnosed as type 1 or type 2 diabetes. Correct diagnosis may result in a change in clinical treatment and impacts prediction of complications and familial risk. In this study, we aimed to assess the prevalence of MODY in multiethnic youth under age 20 years with a clinical diagnosis of type 2 diabetes.
RESEARCH DESIGN AND METHODS
We evaluated whole-exome sequence data of youth with a clinical diagnosis of type 2 diabetes. We considered participants to have MODY if they carried a MODY gene variant classified as likely pathogenic (LP) or pathogenic (P) according to current guidelines.
RESULTS
Of 3,333 participants, 93 (2.8%) carried an LP/P variant in HNF4A (16 participants), GCK (23), HNF1A (44), PDX1 (5), INS (4), and CEL (1). Compared with those with no LP/P variants, youth with MODY had a younger age at diagnosis (12.9 ± 2.5 vs. 13.6 ± 2.3 years, P = 0.002) and lower fasting C-peptide levels (3.0 ± 1.7 vs. 4.7 ± 3.5 ng/mL, P < 0.0001). Youth with MODY were less likely to have hypertension (6.9% vs. 19.5%, P = 0.007) and had higher HDL cholesterol (43.8 vs. 39.7 mg/dL, P = 0.006).
CONCLUSIONS
By comprehensively sequencing the coding regions of all MODY genes, we identified MODY in 2.8% of youth with clinically diagnosed type 2 diabetes; importantly, in 89% (n = 83) the specific diagnosis would have changed clinical management. No clinical criterion reliably separated the two groups. New tools are needed to find ideal criteria for selection of individuals for genetic testing.
Introduction
Monogenic diabetes, including maturity-onset diabetes of the young (MODY), occurs when a single gene abnormality leads to diabetes. Monogenic diabetes can be caused by mutations in genes that disrupt glucose sensing, insulin transcription, the KATP channel that transduces the signal for insulin release, the insulin gene, or pancreatic development. Correct diagnosis of monogenic diabetes has implications for management, prediction of complications, and familial risk (1–5). MODY caused by mutations in hepatocyte nuclear factor (HNF)-1A and HNF-4A is effectively managed with oral sulfonylurea therapy (1,2). MODY due to mutations in glucokinase (GCK) causes a mild, stable hyperglycemia with low risk of complications that commonly does not require any treatment but may have implications for pregnancy management (5). Yet, frequently, the diagnosis of MODY is missed: in 2010, a study in the U.K., Shields et al. (6) examined regional referral patterns and estimated that ∼80% of MODY is not clinically diagnosed by molecular testing. In the U.S., a sequencing study of pancreatic autoantibody-negative, C-peptide–positive participants in the SEARCH for Diabetes in Youth (SEARCH) study found that only 6% of youth found to have pathogenic variants in MODY genes had a prior clinical diagnosis of MODY (7). Sequencing the cohort from the Treatment Options for Type 2 Diabetes in Adolescents and Youth (TODAY) study found 4.5% with genetic variants causing MODY (8). Clinical guidelines frequently cite diabetes occurring in successive generations as a selection criterion for genetic testing (4,9); however, up to half of youth with MODY do not have a parental history of diabetes (7,8). The current trend of a high rate of obesity in youth (10) compounds the difficulty of identifying youth with MODY rather than the increasingly prevalent type 2 diabetes.
The Progress in Diabetes Genetics in Youth (ProDiGY) is a collaborative effort of two pediatric diabetes studies, SEARCH and TODAY, with the Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) consortium. In this study, with whole-exome sequence data from the pediatric participants generated in T2D-GENES, we aimed to assess the prevalence of MODY in youth with a clinical diagnosis of type 2 diabetes. In addition, we examined differences in clinical characteristics between those with and without a monogenic cause of their diabetes.
Research Design and Methods
Participants
ProDiGY is a multiethnic resource that brings together youth with clinician-diagnosed type 2 diabetes before age 20 years from SEARCH (n = 492), participants with type 2 diabetes from the TODAY study (n = 511), and additional case subjects from the ancillary TODAY Genetics study (n = 2,330). A whole-exome sequence from these participants was generated as part of the T2D-GENES consortium. TODAY and SEARCH have previously been described in detail (11–13).
Briefly, SEARCH is a population-based prospective registry launched in 2000 with ascertainment of cases of diabetes in youth diagnosed before 20 years of age in the U.S. Youth with type 2 diabetes were identified by physician report and invited to a study visit. At the study visit, participants underwent physical examination and fasting blood draw. Measurements of diabetes autoantibodies and fasting C-peptide levels were performed, and blood was obtained for DNA analyses.
The TODAY study enrolled participants age 10–17 years, with type 2 diabetes, between 2004 and 2009. Participants were overweight or had obesity (BMI ≥85th percentile for age, sex, and height), with negative pancreatic autoantibodies and a fasting C-peptide level >0.6 ng/mL. Data from physical exams and fasting blood samples performed at the baseline visit were used in this analysis. Of note, American Indian tribal nations elected not to participate in the genomics collection (14). TODAY Genetics is an ancillary study of TODAY in which additional participants with pediatric type 2 diabetes were enrolled outside of the original clinical trial. To qualify, participants must have been diagnosed with type 2 diabetes before 18 years of age and have a documented BMI ≥85th percentile at the time of diagnosis. Exclusion criteria included taking a medication known to affect glucose tolerance, insulin sensitivity, or secretion within 60 days of diagnosis; having a genetic syndrome or disorder known to affect glucose tolerance (other than diabetes); or being a blood relative of a previously enrolled participant. Data and sample collection occurred at a one-time research visit at 1 of 25 clinical sites. During this visit, a self-report questionnaire on family and medical history was administered and blood samples were drawn for DNA extraction and for analysis of glucose, C-peptide levels, and autoantibodies. Although BMI ≥85th percentile at diabetes diagnosis was used as an inclusion criterion, a physical exam, including BMI measurement, was not performed as part of the study and not recorded for analysis.
The TODAY and SEARCH protocols were approved by the institutional review boards of each participating institution, and participants provided written informed parental consent and child assent.
Whole-Exome Sequencing
Genomic DNA was sheared, end repaired, ligated with barcoded Illumina sequencing adapters, amplified, size selected, and subjected to in-solution hybrid capture with the Agilent SureSelect Human All Exon 44Mb v2.0. Resulting Illumina exome sequencing libraries were quantitative PCR quantified, pooled, and sequenced with 76–base pair paired-end reads with Illumina GAII or HiSeq 2000 sequencers to ∼56× mean coverage. Sequence data were processed and aligned to hg19 with use of the Picard (https://broadinstitute.github.io/picard/), Burrows-Wheeler Alignment (BWA) (15), and Genome Analysis Toolkit (GATK) (16,17) pipelines. Of the combined 3,700 youth-onset type 2 diabetes samples, 3,698 samples passed sequencing during variant call set 1, using a passing metric of >80% covered at 20× depth.
Variant Analysis
We focused our analysis on 11 genes previously identified as causal for MODY in the Online Mendelian Inheritance in Man (OMIM) catalog (HNF4A, GCK, HNF1A, PDX1, HNF1B, NEUROD1, CEL, INS, KCNJ11, ABCC8, and APPL1), excluding BLK, PAX4, and KLF11, as these have recently been refuted or disputed as MODY genes by the National Institutes of Health Clinical Genome Resource (ClinGen) (18). Pathogenicity of uncommon (<5% minor allele frequency in ExAC, 1000 Genomes, and Exome Sequencing Project) coding or splicing variants in these genes was classified according to American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines for variant interpretation (19). These guidelines incorporated consideration of computational predictions, clinical phenotypes, functional assessments, and population frequencies and categorized the variants into five categories: (pathogenic [P], likely pathogenic [LP], variant of unknown significance [VUS], likely benign [LB], or benign [B]). We used 10 computational prediction tools in our analysis: Combined Annotation–Dependent Depletion (CADD) (20), Sorting Intolerant From Tolerant (SIFT) (21), PolyPhen-2 (22), MutationTaster2 (23), MutationAssessor (24), likelihood ratio test (LRT) (25), Functional Analysis Through Hidden Markov Models (FATHMM) (26), support vector machine (SVM) (27), logistic regression (LR) (27), and Genomic Evolutionary Rate Profiling (GERP) (28).
Statistical Analysis
We compared clinical characteristics (age at diabetes diagnosis, fasting C-peptide level, family history of diabetes, dyslipidemia, and hypertension) of participants with and without a genetic diagnosis. Dyslipidemia was defined as either self-reported diagnosis of high cholesterol or, as per American Diabetes Association goals for children with diabetes, as LDL ≥100 mg/dL, HDL ≤35 mg/dL, or triglycerides ≥150 mg/dL (29). Hypertension was defined either as self-reported diagnosis of high blood pressure or as per American Diabetes Association goals as systolic or diastolic blood pressure ≥95th percentile for age, sex, and height in children under 13 years or systolic blood pressure ≥130 mmHg or diastolic blood pressure ≥80 mmHg in those age ≥13 years (29). In our primary analysis, we performed comparisons in aggregate (MODY vs. non-MODY). As a secondary analysis, we compared GCK MODY with non-GCK MODY. Continuous measures that were normally distributed were compared by t test, while those not normally distributed were compared by Wilcoxon rank sum test. Categorical values were compared by χ2 test or, where expected counts were too low, by Fisher exact test. We plotted receiver operating characteristic (ROC) curves for those clinical characteristics for which there was a significant statistical difference between participants with and without MODY. An area under the ROC curve (AUC) of >0.8 would be considered indicative of a multivariable model clinically useful for identifying patients for MODY testing. All statistical analysis was performed with STATA 12.1 (StataCorp, College Station, TX).
Results
In both SEARCH and the TODAY Genetics study, pancreatic autoantibodies were measured at baseline, antibody-positive participants were not excluded from genetic sequencing. In the TODAY study pancreatic autoantibodies were measured prior to enrollment, and antibody-positive subjects were not enrolled. After excluding pancreatic autoantibody–positive participants (n = 317 [8.7% of participants]), we had 3,333 youth with type 2 diabetes and whole-exome sequencing data. Age at diagnosis was similar in the three contributing cohorts, averaging 14 years. Sex distribution was also similar, with a female predominance. The cohorts were multiethnic, including individuals of non-Hispanic Black, non-Hispanic White, Hispanic, Asian, and Native American ancestry, though the latter two groups made up a very small proportion of the sample (Table 1). As previously noted, TODAY study participants from Native American tribes elected not to participate in genetic analysis.
Table 1.
Demographics and clinical characteristics of participants (n = 3,333) from the cohorts with youth-onset type 2 diabetes by original study cohort
| TODAY (n = 511) | TODAY Genetics (n = 2,330) | SEARCH (n = 492) | |
|---|---|---|---|
| Age at diagnosis, years, mean (SD) | 13.7 (2.1) | 13.9 (2.3) | 14.1 (2.5) |
| Sex, % male | 36.1 | 34.7 | 38.0 |
| Race/ethnicity, % | |||
| Non-Hispanic White | 20.9 | 18.9 | 23.5 |
| Non-Hispanic Black | 31.4 | 33.3 | 42.9 |
| Hispanic | 43.4 | 40.2 | 28.4 |
| Asian | 1.7 | 2.1 | 2.5 |
| Native American | 0.0 | 0.5 | 0.5 |
| Other/missing | 2.7 | 5.1 | 2.2 |
| Dyslipidemia, % | 61.6 | 17.5 | 71.3 |
| Hypertension, % | 19.2 | 19.6 | 17.4 |
| C-peptide (nmol/L)* | 1.30 | 1.2 | |
| Total cholesterol (mmol/L)* | 3.80 | 4.6 | |
| Triglycerides (mmol/L)* | 1.30 | 1.8 | |
| LDL cholesterol (mmol/L)* | 2.10 | 2.7 | |
| HDL cholesterol (mmol/L)* | 1.00 | 1.1 |
Data are means.
In total, we found 93 participants (2.8%) with a P or an LP variant in a MODY gene. The most common gene identified was HNF1A, with 44 participants (1.3%) carrying a P or an LP variant. Of the other MODY-positive participants, 23 (0.7%) had variants in GCK, 16 (0.5%) in HNF4A, 5 (0.2%) in PDX1, 4 (0.1%) in INS, and 1 in CEL (Table 2 and Supplementary Table 1). MODY variants were identified in all ethnic groups studied except for Native Americans, who represented a very small fraction of the overall sample.
Table 2.
Variants identified in MODY genes
| Gene name | No. of participants with P/LP variants | No. of variants per gene* | No. of novel P/LP variants | No. of variants of unknown significance |
|---|---|---|---|---|
| HNF1A | 44 | 73 | 10 | 41 |
| GCK | 23 | 50 | 2 | 30 |
| HNF4A | 16 | 49 | 9 | 34 |
| PDX1 | 5 | 39 | 2 | 33 |
| INS | 4 | 12 | 1 | 4 |
| CEL | 1 | 87 | 1 | 67 |
| ABCC8 | 0 | 88 | 0 | 86 |
| KCNJ11 | 0 | 33 | 0 | 31 |
| NEUROD1 | 0 | 22 | 0 | 21 |
| APPL1 | 0 | 37 | 0 | 34 |
| BLK | 0 | 49 | 0 | 41 |
Variants per gene includes all variants <5% minor allele frequency, whether classified as B, LB, VUS, LP, or P.
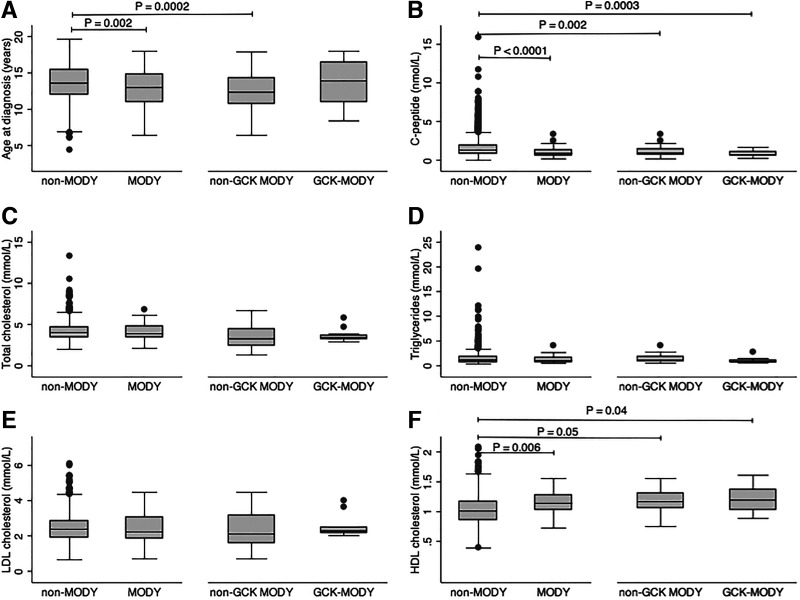
Mean age at diabetes diagnosis was slightly, albeit significantly, lower for those with a P/LP MODY variant (12.9 vs. 13.6 years, P = 0.002), although there was a high degree of overlap in range. This difference appeared to be explained by a lower age at diagnosis for those with non-GCK MODY (12.6 and 13.7 years in non-GCK MODY and GCK MODY, respectively [Fig. 1A]). Fasting C-peptide was significantly lower for those with MODY (1.0 vs. 1.6 nmol/L, P < 0.0001), with similar findings for GCK and non-GCK MODY (Fig. 1B).
Figure 1.
Mean age at diagnosis (A), fasting C-peptide (B), fasting total cholesterol (C), triglycerides (D), LDL cholesterol (E), and HDL cholesterol (F) in non-MODY and MODY participants and in the non-GCK and GCK MODY subgroups.
Youth with MODY had rates of dyslipidemia similar to those of youth without MODY (Table 3), although among the subset with measured lipid data (n = 964), MODY youth had higher HDL cholesterol (1.1 vs. 1.0 mmol/L, P = 0.006). Higher HDL cholesterol was seen in both the GCK and non-GCK MODY groups compared with non-MODY youth (Fig. 1F). There were no significant differences in total cholesterol, LDL cholesterol, or triglycerides between MODY and non-MODY or between the MODY subgroups (Fig. 1C–E). Participants with MODY were less likely to have hypertension than youth without MODY. This finding was similar when we further subdivided the participants with MODY by whether they carried a mutation in GCK (Table 3).
Table 3.
Categorical clinical characteristics in participants with and without a diagnosis of MODY
| All participants | MODY participants | |||
|---|---|---|---|---|
| Non-MODY (n = 3,240) | MODY (n = 93) | Non-GCK (n = 70) | GCK (n = 23) | |
| Male sex | 34.3 | 35.7 | 32.0 | 47.8 |
| Race/ethnicity | ||||
| Non-Hispanic White | 17.7 | 30.1 | 30.0 | 30.4 |
| Non-Hispanic Black | 35.6 | 23.7 | 18.6 | 39.1 |
| Hispanic | 40.0 | 40.9 | 45.7 | 26.1 |
| Asian | 2.2 | 0 | 0 | 0 |
| Native American | 0.4 | 0 | 0 | 0 |
| Other/missing | 0.2 | 1.1 | 1.4 | 0 |
| Dyslipidemia | 32.6 | 28.0 | 30.0 | 21.7 |
| Hypertension | 17.5 | 5.9† | 7.8* | 0 |
| Parent with diabetes | 63.6 | 68.6 | 58.5 | 60.9 |
| Parent, grandparent, or sibling with diabetes | 92.4 | 90.7 | 93.0 | 83.3 |
| Sibling with diabetes | 9.8 | 17.1* | 22.8† | 0 |
Data are percentages.
P < 0.05 compared with non-MODY;
P < 0.01 compared with non-MODY.
In evaluation of ROC curves we found that age at diagnosis (AUC 0.59), C-peptide (AUC 0.67), and HDL (AUC 0.66) could not be used to identify those with MODY. With combination of these characteristics the threshold for clinical utility still was not met (AUC of age at diagnosis, C-peptide, and HDL combined 0.76).
Participants with and without MODY were equally likely to have a parent or grandparent with diabetes, but those with MODY were somewhat more likely to have a sibling with diabetes (Table 3).
The majority of participants with MODY were not receiving indicated treatment. Of those with GCK MODY, only 34.8% were appropriately on no treatment. For those with variants in HNF1A and HNF4A, only 10.3% were on a noninsulin/nonmetformin agent (sulfonylurea in most cases).
Conclusions
In this analysis, we found that 93 of 3,333 youth (2.8%) with clinically diagnosed type 2 diabetes had a P or an LP MODY variant. While we found statistically significant differences for age at diagnosis, C-peptide levels, HDL cholesterol, and presence of hypertension, these differences did not clearly distinguish between MODY and non-MODY diabetes. MODY variants were identified in participants of non-Hispanic White, Black, and Hispanic ancestry. Family history, defined as a parent with diabetes, which is frequently used as a criterion for identifying monogenic diabetes, did not distinguish between the two groups. Moreover, nearly half of all participants found to carry a MODY variant did not have a parent diagnosed with diabetes. For clinicians caring for children with diabetes, our findings imply that a small but substantial fraction of patients with apparent youth-onset type 2 diabetes in fact have MODY.
Our study used whole-exome sequencing data rather than targeted gene sequencing, although for the purposes of this analysis we focused on genes previously identified as causal for MODY. A subset of participants, those from the TODAY study and SEARCH, had previously undergone targeted sequencing analysis for MODY genes; 4.5% (by targeted next-generation sequencing of coding regions of 40 genes including all MODY genes except APPL1 at an average depth of 389×) and 8.0% (by Sanger sequencing of HNF1A, HNF4A, and GCK) were identified to have a P variant in each group, respectively (7,8). In our analysis, we were able to identify all variants previously found by targeted sequencing in these participants, indicating that whole-exome sequencing at a mean depth of coverage of 82× is a viable method for finding MODY variants. Our method did not include noncoding regions, potentially missing pathogenic variants, though relatively few of these have been described, and they are especially challenging to classify. In addition, by following strict ACMG/AMP guidelines for variant classification (19), we may have underestimated the true prevalence of MODY in this sample.
P and LP MODY variants were identified in nearly all ethnic groups in this sample, though the proportion was somewhat higher among non-Hispanic White participants. We suspect that this difference may be due to the dependence of ACMG/AMP variant classification standards on previously published data on genetic variants. Underrepresentation of populations of non-European ancestry is a recognized problem that affects the universal utility of precision medicine. Non-European genomes are less well annotated with a higher rate of variants of unknown significance, which could lead to bias in classification (8,30).
Biomarkers such as C-peptide have the potential to help prioritize patients with diabetes for MODY genetic testing. While in our study we found statistically significant differences in C-peptide and HDL, neither facilitated clear discrimination of MODY from non-MODY diabetes, alone or in combination. Other markers, such as hs-CRP and urinary C-peptide–to–creatinine ratio, have shown promising results in specifically separating the most common types of MODY from type 1 or 2 diabetes. For example, serum hs-CRP level was lower in HNF1A MODY patients than in those with other types of diabetes or control subjects without diabetes. However, these tests are not often performed in the U.S., and previous research proposed various cutoffs, with a lack of uniformity (31). Another potential discriminator arises from the association between HNF4A MODY and increased birth weight related to paradoxical transient neonatal diazoxide-responsive hyperinsulinemic hyperglycemia, which is additive to any impact of maternal diabetes on birth weight (32). No studies have evaluated the utility of birth weight in distinguishing HNF4A MODY, but it has been suggested that finding this phenotype and young-onset diabetes in the same pedigree be considered to increase suspicion of HNF4A MODY (32). In any case, we were unable to assess the utility of birth weight, neonatal hyperinsulinemic hypoglycemia, or biomarkers such as hs-CRP, as these measures were not available for our participants.
With the rise of obesity in recent years in youth overall (10,33–35) and in those with type 1 diabetes (36,37), across race/ethnicity, clinical features of type 1 and type 2 diabetes in youth have become less distinct. One would expect that youth with MODY would follow secular trends in weight gain, making it difficult to discriminate between MODY and type 2 diabetes on the basis of BMI or metabolic abnormalities. Although we did find that hypertension was less common and HDL cholesterol levels were significantly higher in MODY participants in this study, neither of these differences was clinically significant, and these factors cannot be used to distinguish participants with MODY from those without MODY. As individual BMI was not collected in the TODAY Genetics cohort, we could not perform a direct comparison of BMI in our study. One could argue that our inability to distinguish participants with MODY from those with conventional type 2 diabetes stems from the use of overweight/obesity as a selection criterion for a majority of participants in our study. However, in the previously published study of MODY in the SEARCH cohort, where individuals were selected only on the basis of lack of autoantibodies and detectable C-peptide, investigators not find that BMI or metabolic characteristics distinguished participants with MODY at the individual level (7). Similarly, in a British study with widened testing criteria for diagnostic sequencing, investigators identified participants with MODY with clinical features not expected to be present in MODY, including higher BMI and presence of metabolic syndrome (38).
Of 93 youth whom we identified as having MODY, 83 (89%) had a specific genetic diagnosis that would change clinical management: 23 had a GCK variant for which treatment is usually not needed (5) and 60 had variants in HNF1A or HNF4A, which are likely to respond better to sulfonylureas than insulin or other antihyperglycemic medications (1,2). In fact, the TODAY study group found that metformin was ineffective in preserving glycemic control in those with HNF4A MODY (8), with seven of seven failing therapy rapidly; the outcome in those with HNF1A MODY was less clear, with three of five participants failing therapy—a proportion that was not significantly different from that among the overall study population.
The diagnosis of MODY has additional implications for the family members of an identified patient. In our study, while parental history of diabetes did not distinguish between MODY and non-MODY diabetes, rates of parental diabetes were high in both groups, although this information was obtained by questionnaire rather than testing of parents, so parental diabetes may be underreported. We did see higher rates of siblings with diabetes among participants with MODY, which is not unexpected given that siblings of individuals with confirmed MODY carry a 50% risk of MODY themselves. In contrast, pediatric type 2 diabetes is uncommon and in part precipitated by lifestyle risk factors, which may differ between siblings. While individuals with a family history of type 2 diabetes, particularly of early onset, should be aware of their genetic risk (likely polygenic), a molecular diagnosis of MODY enables genetic counseling and possible genetic testing of relatives with diabetes using single-site analysis of the proband’s variant. Such testing, known as cascade testing, makes it possible to further disseminate the treatment selection benefits of a molecular diagnosis, which may in some cases coexist with insulin resistance and type 2 diabetes.
The strengths of our study include a large cohort that included a diverse racial/ethnic mix of participants, comprehensive whole-exome sequence data, and methodical application of ACMG/AMP guidelines. However, a limitation of our approach is that exome sequencing may miss potential nonexonic or splicing causal variants. In addition, ACMG/AMP guidelines frequently classify novel or recently discovered variants as of uncertain significance, particularly missense variants, given the required multiple lines of evidence of causality needed to label a variant as P/LP. We therefore may have missed participants with true MODY. Finally, we did not have measured BMI, blood pressure, or lipid analyses in the TODAY Genetics cohort, which may have affected our ability to identify clinical features distinguishing MODY from non-MODY participants.
The selection criteria used in our study excluded youth who look to have MODY, i.e., antibody-negative youth who are not obese, highlighting that even with this selection, the prevalence of monogenic diabetes remains high. Clinicians face many barriers to ordering genetic testing, including the cost of testing and the uncertainty of reimbursement. Modeling has shown genetic testing for MODY to be cost-effective in current medical practice and potentially cost saving (39,40). As the prevalence of obesity in children has been rising, type 2 diabetes in youth is becoming increasingly common (41). In previous testing of adults it was found that expanding the diagnostic testing criteria doubled the numbers of MODY case participants identified compared with current clinical practice and that the yield was greatest in young adult–onset type 2 diabetes (38). Here we found that a small but appreciable number of children otherwise meeting clinical criteria for type 2 diabetes had evidence of MODY on genetic sequencing; in most cases, the knowledge of this diagnosis would alter management. Thus, regardless of BMI, family history, race/ethnicity, lipid levels, or presence of hypertension, a diagnosis of MODY should be considered in youth-onset diabetes with absence of autoantibodies and evidence of preserved β-cell function.
Article Information
Acknowledgments. SEARCH is indebted to the many youth and their families, and their health care providers, whose participation made this study possible.
This study includes data provided by the Ohio Department of Health, which should not be considered an endorsement of this study or its conclusions.
Funding. The authors acknowledge the involvement of the South Carolina Clinical & Translational Research Institute at the Medical University of South Carolina, National Institutes of Health (NIH)/National Center for Advancing Translational Sciences (NCATS) grants UL1 TR000062 and UL1 Tr001450; Seattle Children’s Hospital and the University of Washington, NIH/NCATS grant UL1 TR00423; University of Colorado Pediatric Clinical and Translational Research Center, NIH/NCATS grant UL1 TR000154; the Barbara Davis Center at the University of Colorado Denver, DERC NIH grant P30 DK57516; the University of Cincinnati, NIH/NCATS grants UL1 TR000077 and UL1 TR001425; and the Children with Medical Handicaps Program managed by the Ohio Department of Health. Sequencing for T2D-GENES cohorts was funded by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant U01DK085526 (Multiethnic Study of Type 2 Diabetes Genes) and National Human Genome Research Institute grant U54HG003067 (Large Scale Sequencing and Analysis of Genomes). The TODAY study was completed with funding from NIDDK and the NIH Office of the Director through grants U01-DK61212, U01-DK61230, U01-DK61239, U01-DK61242, and U01-DK61254; from National Center for Research Resources (NCRR) General Clinical Research Centers Program grants M01-RR00036 (Washington University School of Medicine), M01-RR00043-45 (Children’s Hospital Los Angeles), M01-RR00069 (University of Colorado Denver), M01-RR00084 (Children’s Hospital of Pittsburgh), M01-RR01066 (Massachusetts General Hospital), M01-RR00125 (Yale University), and M01-RR14467 (University of Oklahoma Health Sciences Center); and from NCRR Clinical and Translational Science Awards UL1-RR024134 (Children’s Hospital of Philadelphia), UL1-RR024139 (Yale University), UL1-RR024153 (Children’s Hospital of Pittsburgh), UL1-RR024989 (Case Western Reserve University), UL1-RR024992 (Washington University in St. Louis), UL1-RR025758 (Massachusetts General Hospital), and UL1-RR025780 (University of Colorado Denver). The SEARCH for Diabetes in Youth Cohort Study (SEARCH 4) (1UC4DK108173) is funded by the NIDDK, NIH, and supported by the Centers for Disease Control and Prevention. The Population Based Registry of Diabetes in Youth Study (1U18DP006131, U18DP006133, U18DP006134, U18DP006136, U18DP006138, and U18DP006139) is funded by the Centers for Disease Control and Prevention and supported by the NIDDK, NIH. Sites (SEARCH 1 through 4): Kaiser Permanente Southern California (U18DP006133, U48/CCU919219, U01 DP000246, and U18DP002714), University of Colorado Denver (U18DP006139, U48/CCU819241-3, U01 DP000247, and U18DP000247-06A1), Cincinnati Children’s Hospital Medical Center (U18DP006134, U48/CCU519239, U01 DP000248, and 1U18DP002709), University of North Carolina at Chapel Hill (U18DP006138, U48/CCU419249, U01 DP000254, and U18DP002708), Seattle Children’s Hospital (U18DP006136, U58/CCU019235-4, U01 DP000244, and U18DP002710-01), and Wake Forest University School of Medicine (U18DP006131, U48/CCU919219, U01 DP000250, and 200-2010-35171). J.N.T. was supported by NIDDK K12-DK094721.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the NIDDK.
Duality of Interest. The authors acknowledge the involvement of the Kaiser Permanente Southern California’s Marilyn Owsley Clinical Research Center (funded by Kaiser Foundation Health Plan and supported in part by the Southern California Permanente Medical Group). No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.N.T., J.W.K., and T.I.P. contributed to study conception and design. J.W.K., H.Z., and T.I.P. performed variant analysis. J.N.T. performed statistical analysis of clinical characteristics and drafted the manuscript. S.S., S.E.T., L.L.L., L.E.L.K., J.B.T., F.B., G.I., J.M.L., C.P., J.D., J.F., D.D., and J.C.F. assisted in forming the analysis plan and interpretation of data and reviewed and edited the manuscript. All authors reviewed and approved the manuscript. T.I.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 78th Scientific Sessions of the American Diabetes Association, Orlando, FL, 22–26 June 2018.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.14900994.
References
- 1. Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 2003;362:1275–1281 [DOI] [PubMed] [Google Scholar]
- 2. Pearson ER, Pruhova S, Tack CJ, et al. Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4alpha mutations in a large European collection. Diabetologia 2005;48:878–885 [DOI] [PubMed] [Google Scholar]
- 3. Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT. A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med 2009;26:437–441 [DOI] [PubMed] [Google Scholar]
- 4. Hattersley AT, Greeley SAW, Polak M, et al. ISPAD clinical practice consensus guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes 2018;19(Suppl. 27):47–63 [DOI] [PubMed] [Google Scholar]
- 5. Steele AM, Shields BM, Wensley KJ, Colclough K, Ellard S, Hattersley AT. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA 2014;311:279–286 [DOI] [PubMed] [Google Scholar]
- 6. Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia 2010;53:2504–2508 [DOI] [PubMed] [Google Scholar]
- 7. Pihoker C, Gilliam LK, Ellard S, et al.; SEARCH for Diabetes in Youth Study Group . Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab 2013;98:4055–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kleinberger JW, Copeland KC, Gandica RG, et al. Monogenic diabetes in overweight and obese youth diagnosed with type 2 diabetes: the TODAY clinical trial. Genet Med 2018;20:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. American Diabetes Association . 2. Classification and diagnosis of diabetes: Standards of Care in Diabetes—2017. Diabetes Care 2017;40(Suppl. 1):S11–S24 [DOI] [PubMed] [Google Scholar]
- 10. Skinner AC, Ravanbakht SN, Skelton JA, Perrin EM, Armstrong SC. Prevalence of obesity and severe obesity in US Children, 1999-2016. Pediatrics 2018;141:e20173459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeitler P, Epstein L, Grey M, et al.; TODAY Study Group . Treatment options for type 2 diabetes in adolescents and youth: a study of the comparative efficacy of metformin alone or in combination with rosiglitazone or lifestyle intervention in adolescents with type 2 diabetes. Pediatr Diabetes 2007;8:74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. SEARCH Study Group . SEARCH for Diabetes in Youth: a multicenter study of the prevalence, incidence and classification of diabetes mellitus in youth. Control Clin Trials 2004;25:458–471 [DOI] [PubMed] [Google Scholar]
- 13. Hamman RF, Bell RA, Dabelea D, et al.; SEARCH for Diabetes in Youth Study Group . The SEARCH for Diabetes in Youth study: rationale, findings, and future directions. Diabetes Care 2014;37:3336–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chadwick JQ, Copeland KC, Branam DE, et al. Genomic research and American Indian tribal communities in Oklahoma: learning from past research misconduct and nuilding future trusting partnerships. Am J Epidemiol 2019;188:1206–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laver T, Wakeling M, Knox O, et al. Redefining the pathogenicity of Maturity Onset Diabetes of the Young (MODY) genes: BLK, PAX4 and KLF11 do not cause MODY. In Diabetes UK Professional Conference London, UK, 2018 [Google Scholar]
- 19. Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:Unit7.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361–362 [DOI] [PubMed] [Google Scholar]
- 24. Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol 2007;8:R232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res 2009;19:1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 2013;34:57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 2015;24:2125–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S; NISC Comparative Sequencing Program . Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005;15:901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. American Diabetes Association . 13. Children and adolescents: 2020. Diabetes Care 2020;43(Suppl. 1):S163–S182 [DOI] [PubMed] [Google Scholar]
- 30. Kessler MD, Yerges-Armstrong L, Taub MA, et al.; Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) . Challenges and disparities in the application of personalized genomic medicine to populations with African ancestry. Nat Commun 2016;7:12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang H, Colclough K, Gloyn AL, Pollin TI. Monogenic diabetes: a gateway to precision medicine in diabetes. J Clin Invest 2021;131:e142244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pearson ER, Boj SF, Steele AM, et al. Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. PLoS Med 2007;4:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hales CM, Carroll MD, Fryar CD, Ogden CL. Prevalence of obesity among adults and youth: United States, 2015-2016. NCHS Data Brief 2017;288:1–8 [PubMed] [Google Scholar]
- 34. Hales CM, Fryar CD, Carroll MD, Freedman DS, Ogden CL. Trends in obesity and severe obesity prevalence in US youth and adults by sex and age, 2007-2008 to 2015-2016. JAMA 2018;319:1723–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wijnhoven TM, van Raaij JM, Spinelli A, et al. WHO European Childhood Obesity Surveillance Initiative 2008: weight, height and body mass index in 6-9-year-old children. Pediatr Obes 2013;8:79–97 [DOI] [PubMed] [Google Scholar]
- 36. Liu LL, Lawrence JM, Davis C, et al.; SEARCH for Diabetes in Youth Study Group . Prevalence of overweight and obesity in youth with diabetes in USA: the SEARCH for Diabetes in Youth study. Pediatr Diabetes 2010;11:4–11 [DOI] [PubMed] [Google Scholar]
- 37. van Vliet M, Van der Heyden JC, Diamant M, et al. Overweight is highly prevalent in children with type 1 diabetes and associates with cardiometabolic risk. J Pediatr 2010;156:923–929 [DOI] [PubMed] [Google Scholar]
- 38. Thanabalasingham G, Pal A, Selwood MP, et al. Systematic assessment of etiology in adults with a clinical diagnosis of young-onset type 2 diabetes is a successful strategy for identifying maturity-onset diabetes of the young. Diabetes Care 2012;35:1206–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naylor RN, John PM, Winn AN, et al. Cost-effectiveness of MODY genetic testing: translating genomic advances into practical health applications. Diabetes Care 2014;37:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peters JL, Anderson R, Shields B, et al. Strategies to identify individuals with monogenic diabetes: results of an economic evaluation. BMJ Open 2020;10:e034716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dabelea D, Mayer-Davis EJ, Saydah S, et al.; SEARCH for Diabetes in Youth Study . Prevalence of type 1 and type 2 diabetes among children and adolescents from 2001 to 2009. JAMA 2014;311:1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]