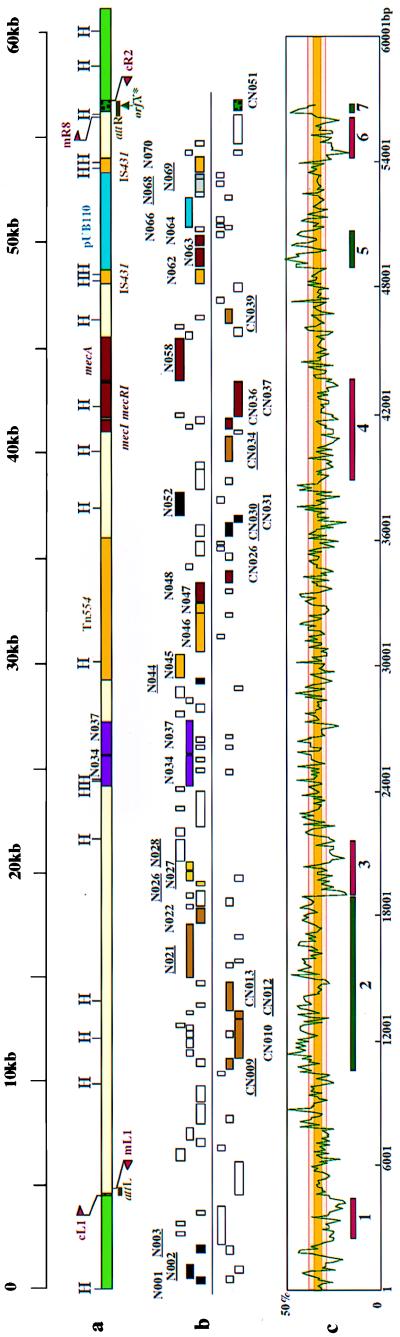
FIG. 2.
(a) The essential structure of mec DNA. Locations of the essential genes are illustrated. Only restriction sites of HindIII are indicated. attL and attR (tentatively defined by the primer pairs cL1 with mL1 and mR8 with cR2) are shown by bars. orfX* is indicated by an arrow. (b) ORFs in and around mec DNA. The ORFs more than 200 bases in size in six possible reading frames are indicated by squares. Those above and those below the line are the ORFs whose transcription is directed to the right and those whose transcription is directed to the left, respectively. Colored squares indicate the ORFs whose extant gene homologues were found in the databases, though many of them were considered incomplete ORFs (underlined). These are potentially defective genes or pseudogenes containing either deletion, nonsense mutation, or frameshift mutation. The colors correspond to the inferred functional properties of the ORFs: red, drug resistance; yellow, transposases (for Tn554, IS431, and putative transposons); gray, plasmid replication (rep for pUB110); magenta, site-specific recombinase (N034 and N037); brown, metabolism and physiology; blue, plasmid replication (for pUB110); black, unknown function. CN051 (stippled green) corresponds to orfX*. Three ORFs, N001 to N003, located outside mec DNA have unknown function (black). (c) Regional change of G+C content in and around mec DNA. The G+C content was calculated and plotted as a line graph by using the Window program of The Wisconsin Package, with a window size of 100 bases and a shift increment of 100. The yellow bar indicates the range of G+C content of S. aureus strains, which is 32 to 36% (34). The range delimited by the orange bars is that of the genus Staphylococcus, which is 30 to 39% (34). The regions with substantially lower than average G+C content (regions 1, 3, 4, 6) and with high G+C content (regions 2, 5, 7) are indicated by red bars and green bars, respectively.