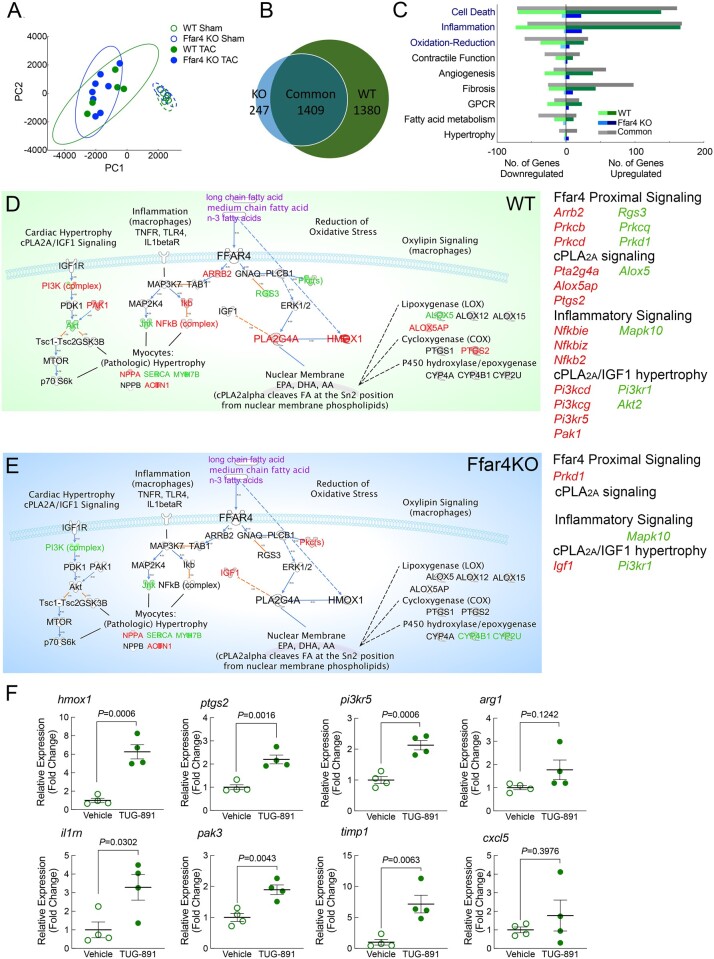
Figure 2.
Transcriptome analysis (RNA-seq) was performed on cardiac myocytes isolated from male WT and Ffar4KO mice 3-days post-TAC or sham surgery. (A) Principal component analysis of RNA transcriptomes from WT and Ffar4KO cardiac myocytes. (B) Venn diagram indicating genes differentially expressed ≥ 1.7-fold uniquely in WT cardiac myocytes post-TAC (1380 genes), upregulated ≥ 1.7-fold uniquely in Ffar4KO cardiac myocytes post-TAC (247 genes), or upregulated ≥ 1.7-fold in both (1409). (C) Differentially expressed genes identified in B were sorted based on gene ontology (GO) terms for biological function. Graphical representation of the number of genes up- or down-regulated in each category that were unique to the WT TAC (relative to sham, green), unique to Ffar4KO TAC (relative to sham, blue), and shared between WT and Ffar4KO (grey). (D, E) Data sets of gene expression from WT (D) and Ffar4KO (E) cardiac myocytes were analysed using Ingenuity Pathway Analysis Software (version 01-16). A custom pathway for known Ffar4 signalling pathways was generated, and expression data for WT and Ffar4KO were analysed (WT, green map, Ffar4KO blue map, as indicated). Genes upregulated (red) or down-regulated (green) for each group are indicated. (F) Induction of gene cardiac gene expression by TUG-891 (35 mg/kg/d, IP, for 3 days) in male WT mice. Each gene is indicated at the top of the graph. Data are mean ± SEM, n = 4 mice per group, data were analysed by Student’s t-test.