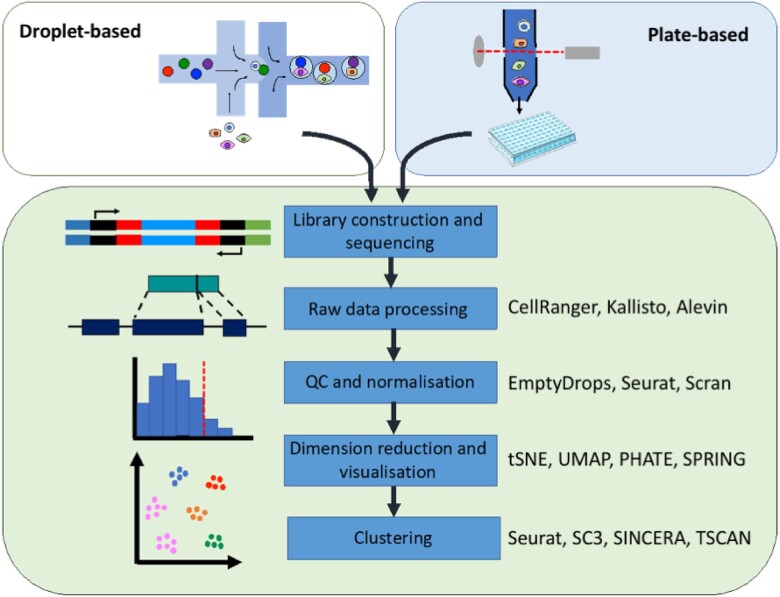
Figure 2.
Schematic of a typical scRNA-seq workflow: single cells are separated and sequencing libraries generated using either droplet or plate-based scRNA-seq platforms. Raw sequencing data are processed and aligned to the reference genome to generate count matrices. Following quality control (QC) and normalisation, dimension reduction is performed to facilitate visualisation in low dimensional space. Subsequent clustering is performed to group cells by their similarity in transcriptional profile. Frequently used bioinformatic tools for each of these steps are indicated on the right side.