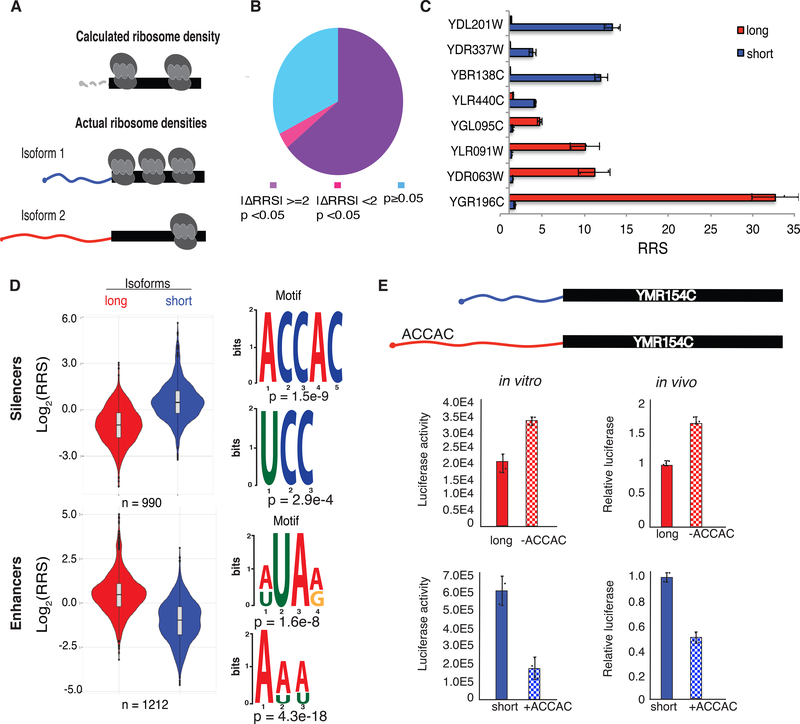
Fig. 4. DART identifies translational silencers and enhancers within alternative 5′ UTRs.
(A) 5′ UTR isoforms confound the interpretation of averaged ribosome densities observed in coding sequences by ribosome profiling. (B) Most alternative 5′ UTR isoforms tested affect ribosome recruitment (RRS). 2653 soform pairs differed significantly by >2-fold (purple), 145 differed significantly, but by <2-fold (magenta), and 1332 did not confer significant differences in RRS (blue). P values from t test Bonferroni corrected. (C) Example genes with alternative 5′ UTR isoforms differing in RRS. The long (5′ extended) isoforms of YDL201W, YDR337W, YBR138C and YLR440C contain putative silencer elements and the long isoforms of YGL095C, YLR091W, YDR063W and YGR196C contain putative enhancers. (D) Violin plots of RRS values from isoform pairs showing differential ribosome recruitment (p < 0.05). The additional 5′ sequences included in longer isoforms were used as inputs to identify enriched motifs using DREME. Top two silencing (above) and enhancing (below) motifs are shown. (E) ACCAC motifs identified by DART analysis are sufficient to repress translation of full-length mRNAs in vitro (left) and in vivo (right). Translation activity in vivo is normalized to Fluc mRNA levels and total protein. See also Supplementary Fig. 3 and 4.