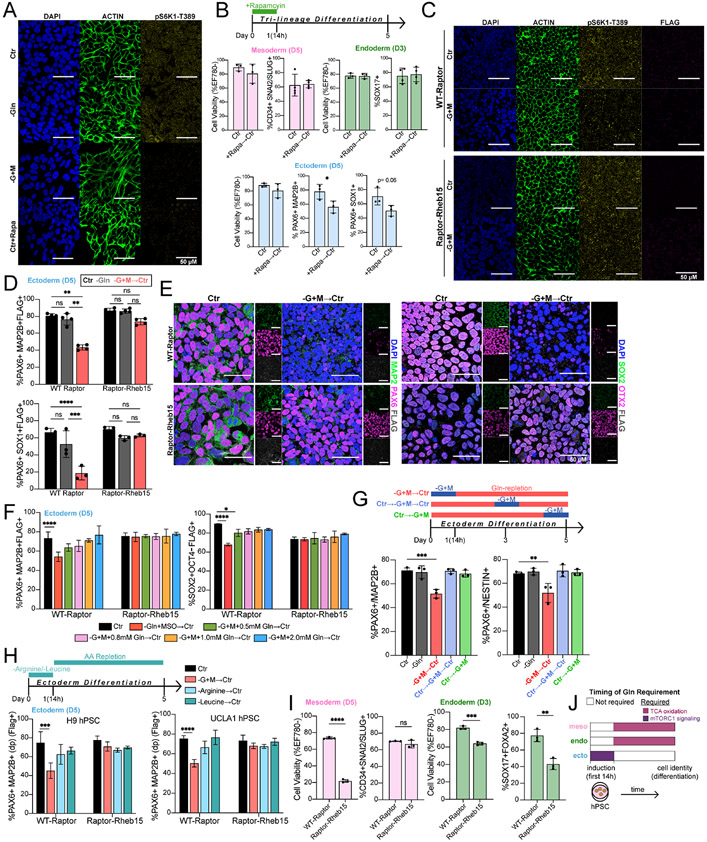
Figure 4. Ectoderm differentiation requires initial Gln-dependent mTORC1 signaling.
(A) Representative immunofluorescence images of mTORC activation in H9-derived ectoderm cells grown for 24h in Gln-supplemented (Ctr), Gln-free (−Gln), Gln-starvation (−G+M), or 200 nM rapamycin treatment (Ctr+Rapa). DAPI (blue, nucleus); ACTIN (green, cytoskeleton); pS6K1-Thr389 (yellow, mTORC1 activation). Scale bar indicates 50 μm.
(B) Percentage of H9-derived D5 mesoderm (CD34+SNAI2/SLUG+), D3 endoderm (SOX17+FOXA2+), and D5 ectoderm (PAX6+MAP2B+, PAX6+SOX1+) cells after initial 14h 200 nM rapamycin treatment.
(C) Representative immunofluorescence images of mTORC activation in H9-derived WT-Raptor or Raptor-Rheb15 ectoderm cells grown for 12h in Gln-supplemented (Ctr) or Gln-starvation (−G+M). DAPI (blue, nucleus); ACTIN (green, cytoskeleton); pS6K1-Thr389 (yellow, mTORC1 activation); FLAG tag (magenta; transduced cell line reporter). Scale bar indicates 50 μm.
(D) Percentage of H9-derived D5 WT-Raptor or Raptor-Rheb15 ectoderm (PAX6+MAP2B+FLAG+, PAX6+SOX1+FLAG+) cells after initial 14h Gln-starvation (−G+M→Ctr).
(E) Representative immunofluorescence images of ectoderm biomarkers in H9-derived D5 (Top) WT-Raptor or (Bottom) Raptor-Rheb15 ectoderm cells grown in Gln-supplemented (Ctr) or Gln-repletion follow initial 14h Gln-starvation (−G+M→Ctr). (Left) MAP2B (green, ectoderm;) PAX6 (magenta, ectoderm); FLAG (grey, transduced cell line reporter). (Bottom) SOX2 (green, ectoderm/pluripotent); OTX2 (magenta, ectoderm), FLAG (grey, transduced cell line reporter). Scale bar indicates 50 μm.
(F) Percentage of ectoderm (PAX6+MAP2B+FLAG+) and pluripotent (SOX2− OCT4+FLAG+) biomarkers in H9-derived D5 WT-Raptor or Raptor-Rheb15 ectoderm cells after initial 14h Gln-starvation with increasing concentrations of exogenous Gln supplementation.
(G) Percentage of H9-derived D5 ectoderm (PAX6+MAP2B+, PAX6+NESTIN+) cells after initial (−G+M→Ctr), intermediate (Ctr→−G+M→Ctr), or late (Ctr→−G+M) 14h Gln-starvation.
(H) Arginine (Arg) or Leucine (Leu) were removed from culture media for 14h then switched to Arg/Leu-supplemented conditions (AA repletion) until D5. Percentage of (Left) H9 or (Right) UCLA1-derived D5 WT-Raptor or Raptor-Rheb15 ectoderm (PAX6+MAP2B+FLAG+) cells after initial 14h Gln-starvation, Arg deprivation, or Leu deprivation.
(I) Cell viability and percentage of H9-derived WT-Raptor or Raptor-Rheb15 D5 mesoderm (CD34+SNAI2/SLUG+FLAG+) and D3 endoderm (SOX17+FOXA2+FLAG+) cells.
(J) Timing of Gln requirement for downstream lineage cell identity. Gln is required as a metabolite precursor in mesoderm and endoderm cells and as a mTORC1 signaling activator in ectoderm cells. Related to Figure 3K.
Data represent mean ± SD of n ≥ 3 biological replicates. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. The p values were determined by (B, I) unpaired two-tailed Student’s t test, (D, F, H) two-way ANOVA, or (G) one-way ANOVA with correction for multiple comparisons.