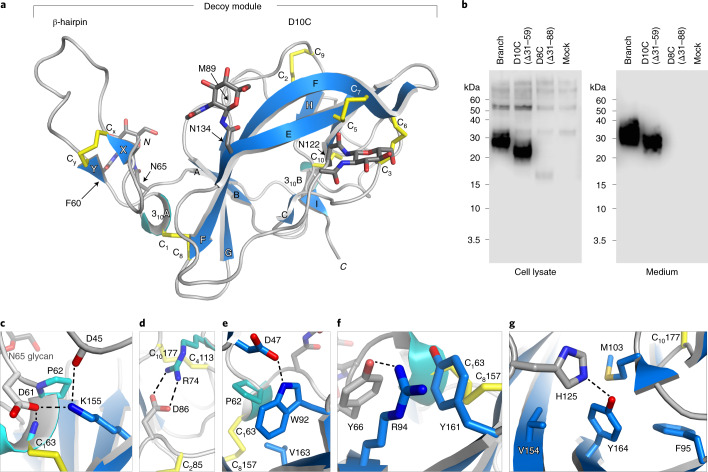
Fig. 1. The GP2 branch region includes a D10C domain whose new fold explains patient mutations in UMOD.
a, Overall structure of the GP2 branch region/decoy module, depicted in cartoon representation with β-strands in blue, 310 helices in cyan and loops in light gray. Disulfides and glycans are shown as yellow and dark gray sticks, respectively, with oxygen atoms in red and nitrogen atoms in blue. b, Reducing western blot comparison of the expression and secretion of GP2 constructs corresponding to the entire branch, D10C or D8C. n = 3. c–g, Details of the GP2 structure rationalize the effect of kidney disease-associated UMOD mutations affecting a set of residues identical between the two proteins (Supplementary Table 2). Selected GP2 D10C domain residues and mutations affecting the corresponding identical residues of UMOD are as follows: GP2 D61, P62, C163→UMOD D172H, P173L/R, C174R (c); GP2 R74, C285, D86, C4113, C10177→UMOD R185C/G/H/L/S, C195F/Y, D196N/Y, C223R/Y, C287F (d); GP2 P62, C163, W92, C8157, V163→UMOD P173L/R, C174R, W202C/S, C267F, V273F/L (e); GP2 C163, R94, C8157→UMOD C174R, R204G/P, C267F (f); GP2 Y164, C10177→UMOD Y274C/H, C287F (g).