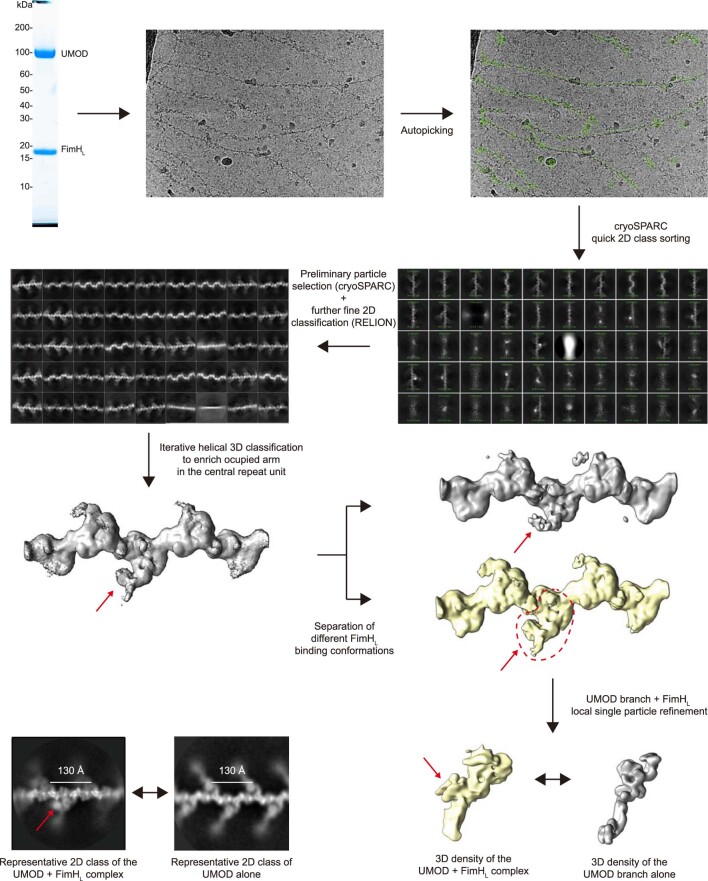
Extended Data Fig. 9. 3D reconstruction of the UMOD branch/FimHL complex.
Identification, isolation and local refinement of a single UMOD branch unit bound to one copy of FimHL. After incubation with an excess concentration of FimHL, UMOD filaments were subjected to cryo-EM analysis. Following filament autopicking by an in-house script, highly heterogenous filament segments were sorted by performing cryoSPARC 2D class runs, after binning. Segment coordinates from good 2D classes were then extracted and re-imported into RELION. After iterative 3D classification with and without applying helical symmetry, the segments with higher FimHL occupancy were selected and grouped into different sub-classes. Segments representing a single branch unit of the best UMOD/FimHL sub-class were extracted and used for 3D reconstruction of the density of UMOD bound to FimHL. In the bottom left panel, the extra density of FimHL in the UMOD branch/FimHL complex could be identified in the 2D class images. Red arrows point to the location of FimHL.