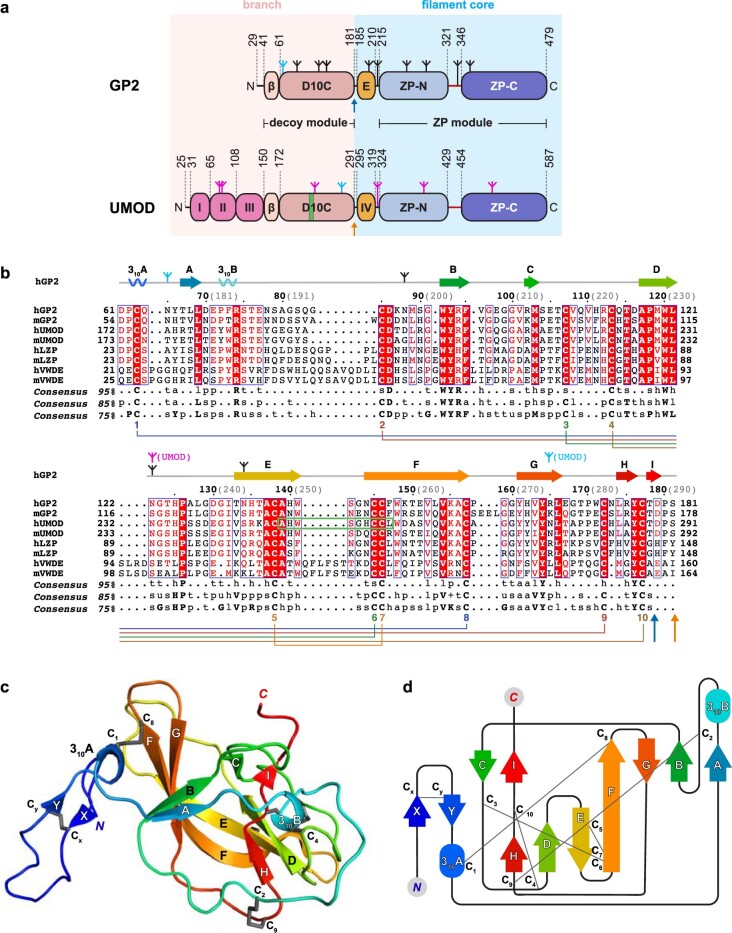
Extended Data Fig. 1. Structure of the GP2 N-terminal branch and its relation with the corresponding regions of UMOD and additional mammalian proteins.
a, Domain architecture of mature human GP2 and UMOD. Domains are indicated by their acronyms, except for UMOD epidermal growth factor (EGF) domains that are labeled according to their roman number, the single EGF domain of GP2 (corresponding to UMOD EGF IV) that is labeled as ‘E’ and the β-hairpin of the decoy module (‘β’). The UMOD D10C epitope recognized by Bence-Jones proteins (BJP)14 is shown as a green stripe. Black and magenta inverted tripods indicate the N-glycosylation sites of GP2 and UMOD, respectively, with the high-mannose chains attached to GP2 N65 (this study) and UMOD N2758,12 colored cyan. The position corresponding to the alternative 3’ splice site generating the β isoform of GP2 (T178 | D179)61 and the elastase cleavage site of UMOD (S291 | S292)62 are indicated by vertical blue and orange arrows, respectively. b, Alignment of D10C domain sequences from human (h) and murine (m) homologues of GP2 and UMOD, as well as liver-specific zona pellucida protein (LZP/OIT3, a molecule that can also interact with UMOD in the kidney and urine63) and von Willebrand factor D and EGF domain-containing protein (VWDE; a protein involved in appendage regeneration in a variety of vertebrate species64). Identical residues are highlighted in white and shaded in red; conserved residues are red and marked by blue frames when clustered. Consensuses at different sequence identity thresholds, based on a comprehensive alignment of homologous sequences, are also reported (bold uppercase characters: amino acids with the same one-letter code; regular lowercase characters: l, [I,V,L]; h, [F,Y,W,H,I,V,L]; + , [H,K,R]; -, [D,E]; p, [Q,N,S,T,C,H,K,R,D,E]; u, [G,A,S]; s, [G,A,S,V,T,D,N,P,C]; t, [G,A,S,Q,N,S,T,C,H,K,R, D,E]; (.), any amino acid). GP2 secondary structure elements, rainbow-colored from blue (N-terminus) to red (C-terminus), and disulfide bond connectivity are shown above and below the alignment, respectively. Other elements are labeled as in (a), with a green box indicating the BJP epitope14. Black bold numbers above the alignment indicate hGP2 residues; light grey numbers between parentheses refer to the corresponding hUMOD residues. c, Cartoon representation of the GP2 decoy module, rainbow-colored following the same scheme used for the secondary structure elements of (b). Disulfide bonds are represented as grey sticks. d, Topology and disulfide connectivity diagram of the decoy module.