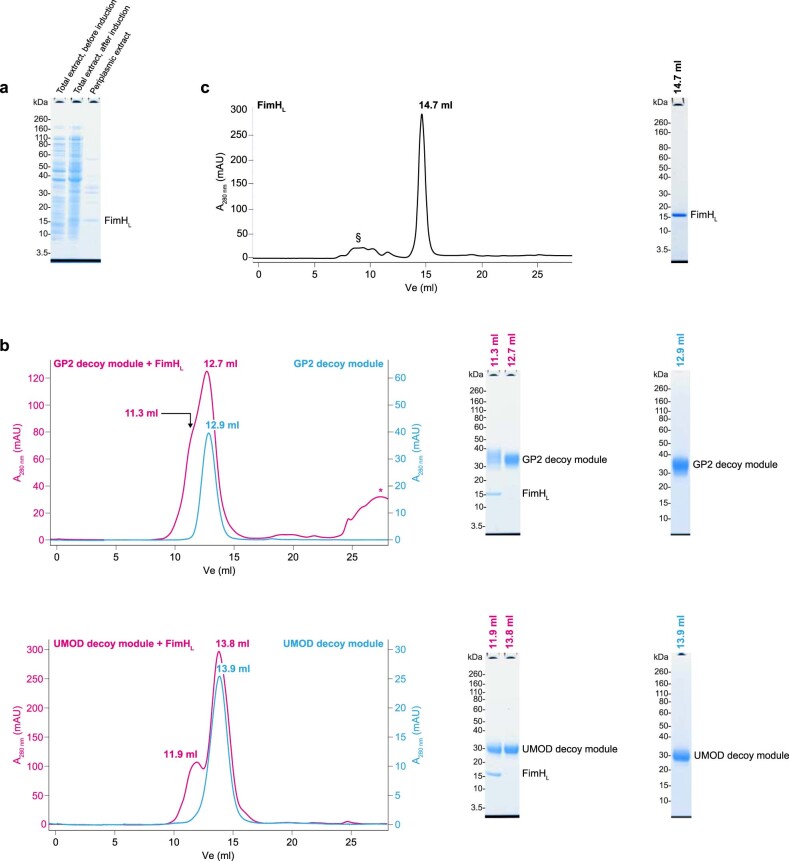
Extended Data Fig. 2. The isolated GP2 branch and the corresponding decoy module of UMOD bind FimHL.
a, For assessing whether the lectin domain of FimH is able to bind in vitro to the branch of GP2 or the equivalent region of UMOD (corresponding to the respective decoy modules, see main text), untagged FimHL was expressed in E. coli and a crude periplasmic extract was prepared. n = 2. b, SEC analysis of the material eluted after incubating purified His-tagged GP2 or UMOD decoy modules bound to IMAC beads with the FimHL-containing E. coli periplasmic extract (magenta curves). In both cases, reducing SDS-PAGE of peak fractions and tandem mass spectrometry (MS/MS) of the corresponding ~15 kDa bands show the presence of complexes between the decoy modules and the bacterial adhesin, indicating that the former are able to selectively recognize the latter among the pool of periplasmic proteins. SEC elution profiles of the GP2 and UMOD decoy domains by themselves are also shown (light blue curves), and a low-molecular weight contaminant peak is indicated by *. GP2 decoy module, UMOD decoy module: n = 3; GP2 decoy module/FimHL, UMOD decoy module/FimHL, n = 2. c, Control SEC profile of unbound His-tagged FimHL with SDS-PAGE analysis of the peak. § indicates minor high-molecular weight contaminants eluting with or close to the void volume. n = 3.