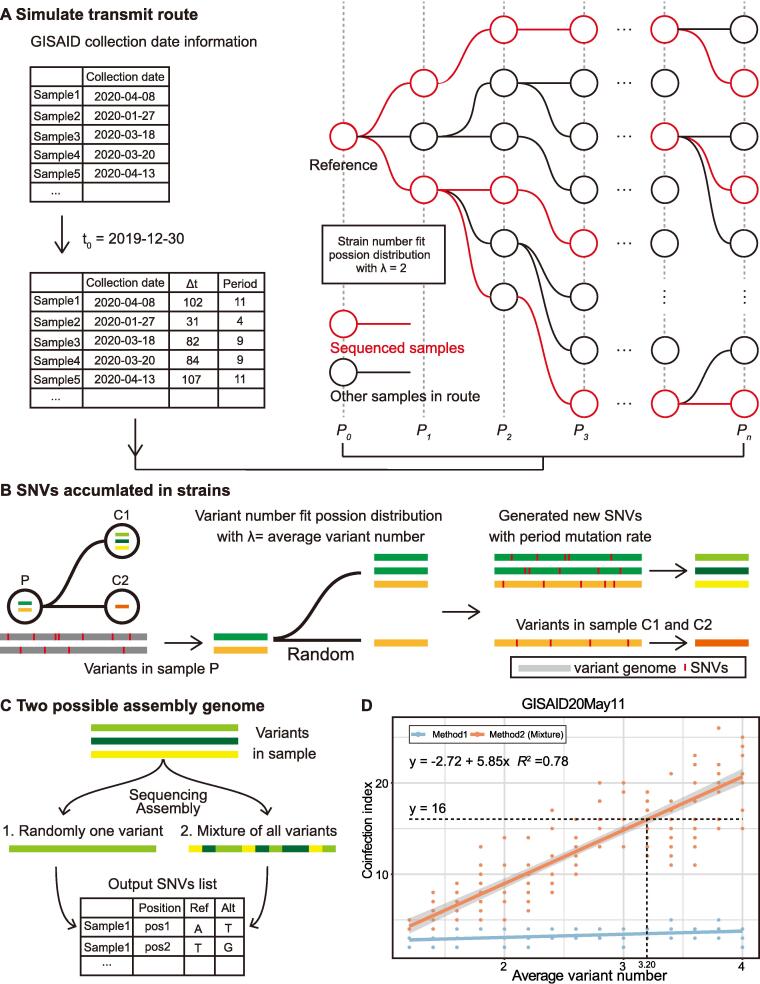
Fig. 3.
The simulation flowchart of viral SNVs in samples and the regression of variant number and the coinfection index. A. We simulated the transmission route based on known epidemiological information of SARS-CoV-2 and constructed a transmission tree. Then, we selected the sequenced samples based on their release date in the GISAID database. B. Variant numbers in all samples fit the Poisson distribution with λ equal to the average variant number. In a single transmission branch, variants in child nodes are randomly inherited from the parent sample. In addition, new SNVs, single-nucleotide variants, would also emerge in the child nodes based on the given mutation rate. C. We simulated two possible methods to obtain assembled genomes in the datasets. In method 1, the assembled genome exhibited one of the variants. In method 2, the assembled genome represented the mixed genome of multiple variants. Then, we acquired the SNV list for all samples as the output. D. The distribution of the coinfection index with different average variant numbers in the GISAID20May11 dataset. The dash lines suggested the coinfected variant number corresponding to the 16-SNP clique with method 2.