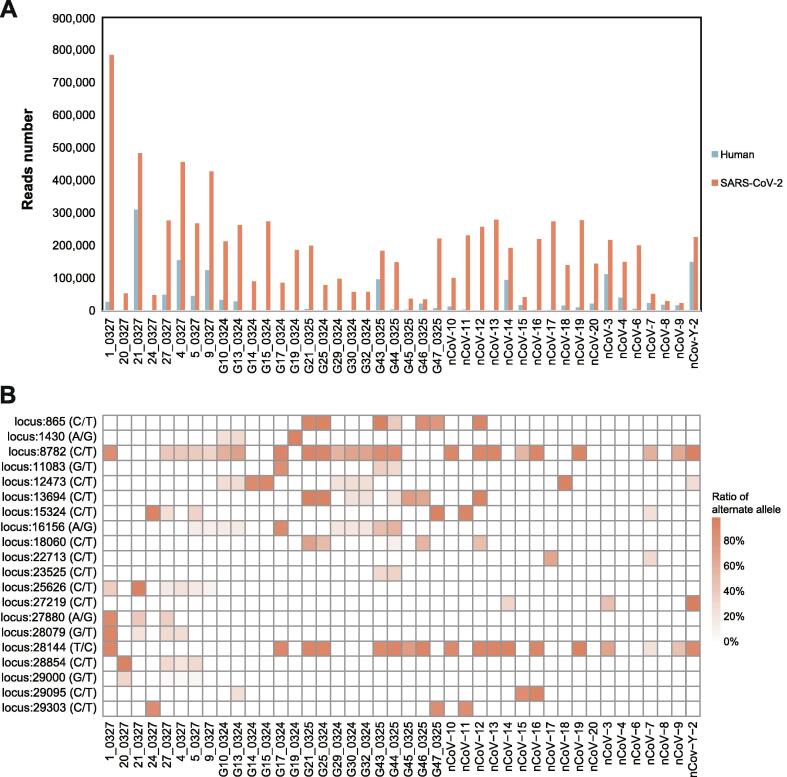
Fig. 6.
Statistics of Nanopore sequencing data and the distribution of recurrent SNPs for the 42 COVID-19 samples. A. After low-quality filtration, we aligned the Nanopore sequencing reads to the SARS-CoV-2 genome and human transcriptome. The histograms in red and blue exhibit the number of reads aligned to the SARS-CoV-2 genome and human transcriptome, respectively. B. The heatmap exhibited twenty recurrent SNPs for the 42 COVID-19 patients. The squares with deeper colour represent the higher ratios of the alternate allele in the sample. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)