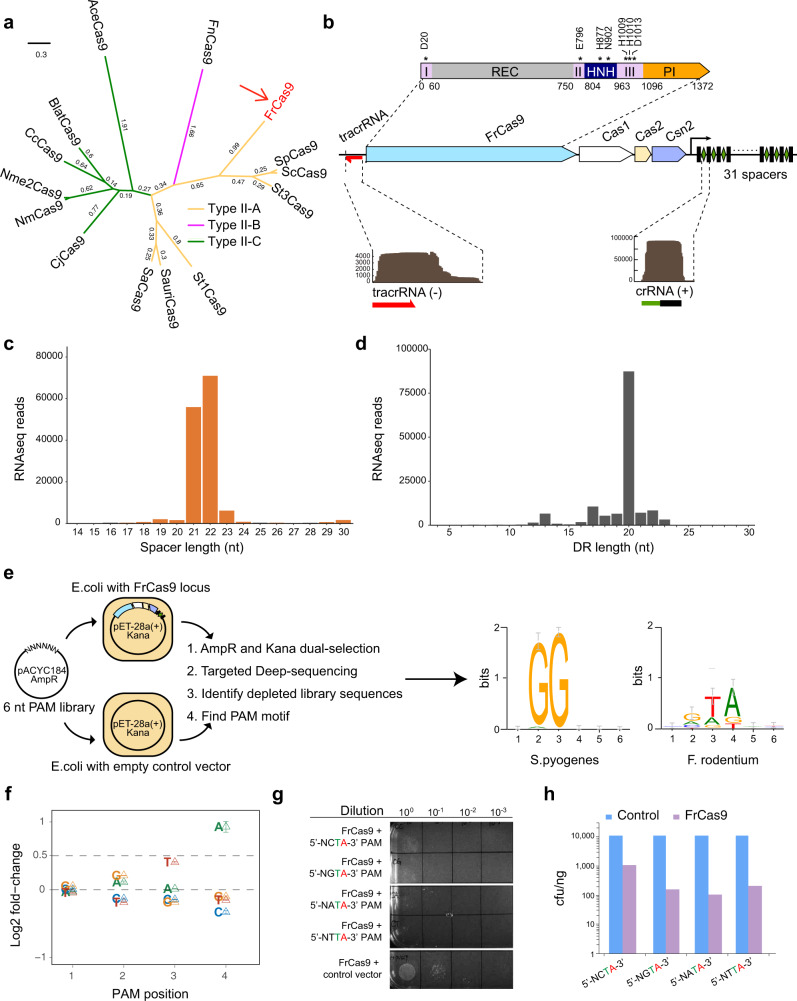
Fig. 1. FrCas9 edits distinct 5′-NNTA-3′ PAMs.
a The phylogenetic tree of FrCas9 and 13 active Cas9 orthologs. b The schematic of Faecalibaculum rodentium systems. Insert above displayed the domains of FrCas9 with active residues indicated with asterisks. I, II, III represented three RuvC domains. Insert below showed expressed crRNA and tracrRNA from small RNA-seq of E. coli harboring pET-28A plasmid with simplified FrCas9 locus. c Distribution of the length of spacer sequences derived from small RNA sequencing. d Distribution of the length of DR sequences derived from small RNA sequencing. e The schematic of depletion assay and web-logo results for SpCas9 and FrCas9. f The deletion effects of first four nucleotides of FrCas9 PAM. Data are presented as mean values ± S.E.M. (n = 4). g The plasmid interference assay of FrCas9 in 4 sites that differed in the 2nd PAM positions. A series of dilution was performed. h The bar plot of cell units of above plasmid interference assay. Source data are provided with this paper.