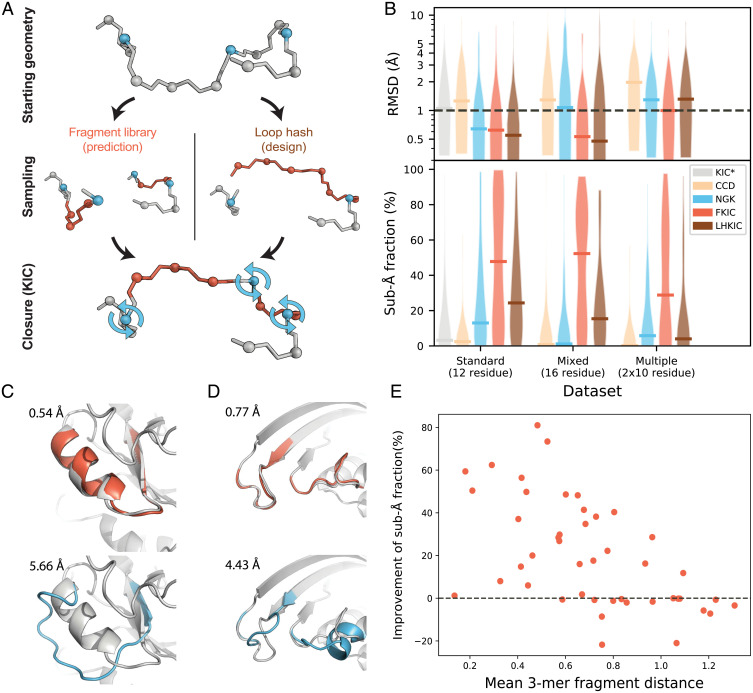
Fig. 2.
FKIC improves prediction of conformations of local backbone segments. (A) Individual FKIC/LHKIC move. Three Cα atoms (blue) on the target segment to be modeled (gray) are picked randomly as pivots. Fragment insertion (FKIC) or loop hash (LHKIC) is applied to sample torsion degrees of freedom at nonpivot atoms (red), which breaks the chain. The KIC algorithm is then used to close the chain by determining appropriate values for the pivot torsions. (B) Comparison of performance of different methods for three datasets: 1) the Standard dataset described in ref. 17 and two new sets, 2) a “Mixed Segment” dataset with 30 16-residue regions that contain both loops and segments of regular secondary structure and 3) a “Multiple Segments'' dataset of 30 cases with two separate 10-residue regions that are interacting. KIC (17): gray; CCD (24): orange; NGK (26): blue; FKIC: red; LHKIC: brown. Upper: violin plot of RMSD of lowest energy (best) model across each dataset. Horizontal bars indicate the median lowest-energy RMSD. FKIC is the only method that provides predictions with atomic accuracy (≤1 Å median RMSD) for all datasets. Lower: violin plot of fraction of predicted models in each dataset that have subångstrom accuracy. FKIC leads to considerable improvements over previous methods. Asterisk indicates data from ref. 42; all other simulations were run with the ref2015 Rosetta energy function (21); methods using fragments (CCD and FKIC) used identical fragment libraries that excluded fragments from structural homologs to the target proteins. (C and D) FKIC accurately predicts geometries from sequence in which the previous state-of-the-art method, NGK, fails. Shown are examples from the Mixed Segment (C) and Multiple Segments dataset (D). Experimentally determined structures: gray; predictions from FKIC: red, Top; predictions from NGK: blue, Bottom. RMSDs to the experimentally determined structures are given in each panel in Å. (E) The fraction of subångstrom predictions is negatively correlated with the mean 3-mer fragment distance (Methods). Each data point represents a protein from the Standard 12-residue dataset.