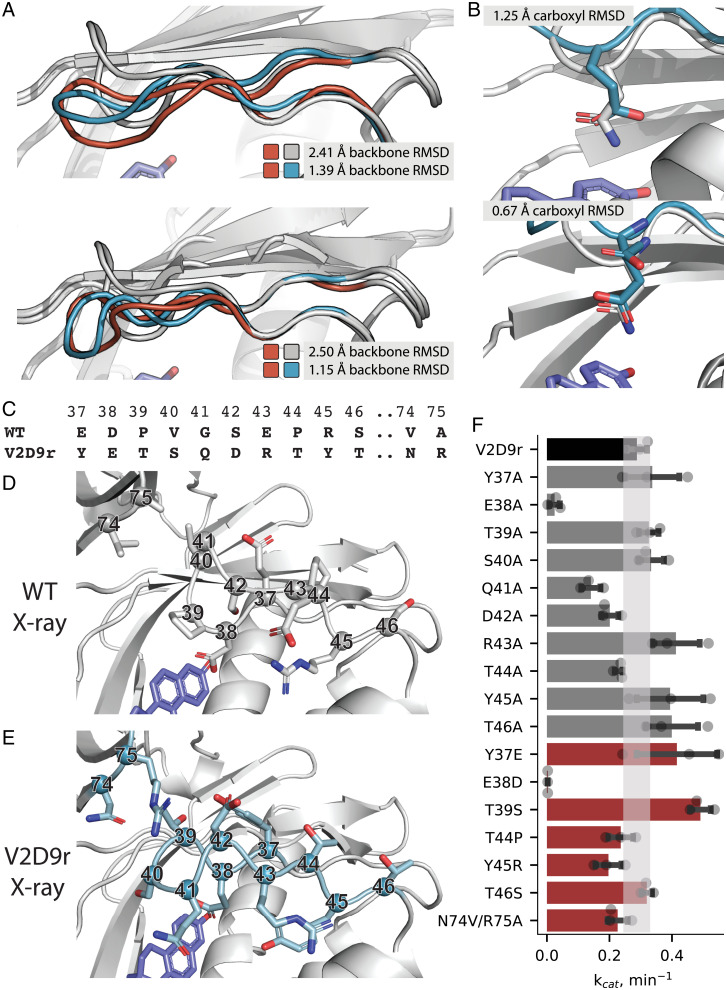
Fig. 4.
Structural characterization of designs V1D8r and V2D9r. (A) Overlay of wild-type KSI crystal structure (gray), lowest-energy predicted models for V1D8r (orange, Top) and V2D9r (orange, Bottom), and crystal structures for V1D8r (blue, Top) and V2D9r (blue, Bottom). (B) Crystal structure (blue) of V1D8r (Top) and V2D9r (Bottom) showing the catalytic glutamate’s placement relative to the amide in the KSI starting structure (PDB 1QJG) used to define the catalytic position (gray). RMSD values between compared structures are indicated in the different panels. (C–F) Mutational analysis of differences between wild-type KSI and design V2D9r: sequence alignment (C), comparison between the active site region in the crystal structures of wild-type KSI (D) and in design V2D9r (E), and (F) bar graph of kcat values for design V2D9r (black), alanine scan mutants (gray), and reversion/selected mutants (red). In F, SDs of independent triplicate experiments are shown as error bars with individual measurements shown as points. The kcat error range for V2D9r is shown as a shaded bar.