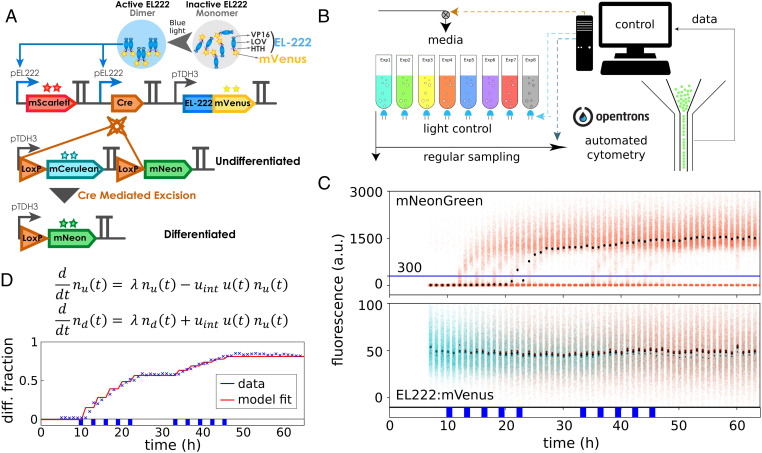
Fig. 1.
A yeast optogenetic differentiation system. (A) EL222:mVenus is constitutively produced from a pTDH3 promoter. In the dark, the LOV domain binds the HTH domain and inhibits dimerization and DNA binding. Upon exposure of cells to blue light, this inhibition is disrupted, and EL222:mVenus dimerizes, binds to the pEL222 promoter, and triggers the expression of mScarlet-I and Cre-recombinase. Cre excises a DNA fragment placed between two target LoxP sites that is designed such that cells switch from producing mCerulean to mNeonGreen from a second pTDH3 promoter. (B) Yeast cells harboring the differentiation system are grown in parallel, fully automated bioreactors that are equipped with controllable blue LEDs. Average optical density of cultures is maintained constant, and flow cytometry measurements are taken at regular intervals via a custom-programmed pipetting robot. (C) Upon exposure of the population to short light pulses, some cells recombine as indicated by increasing mNeonGreen levels in a part of the population (Top). Cells are classified as differentiated when they exceed 300 arbitrary units (a.u.) mNeonGreen fluorescence after deconvolution of signals from the four fluorescent proteins (blue line; SI Appendix, section S9). EL222:mVenus is constitutively expressed in all cells, and there are no clearly noticeable differences in EL222:mVenus levels between differentiated and undifferentiated cells (Bottom). Dots show median fluorescence (black, all cells; cyan, undifferentiated cells; orange, differentiated cells). The applied light sequence is shown at the bottom (two times five 1-h pulses with 2 h between subsequent pulses). The data of all four fluorescent reporters are visualized as fluorescence histograms in SI Appendix, Fig. S1. (D) A simple model is capable of capturing population dynamics of differentiated and undifferentiated cells very well. The applied light sequence is shown at the bottom.