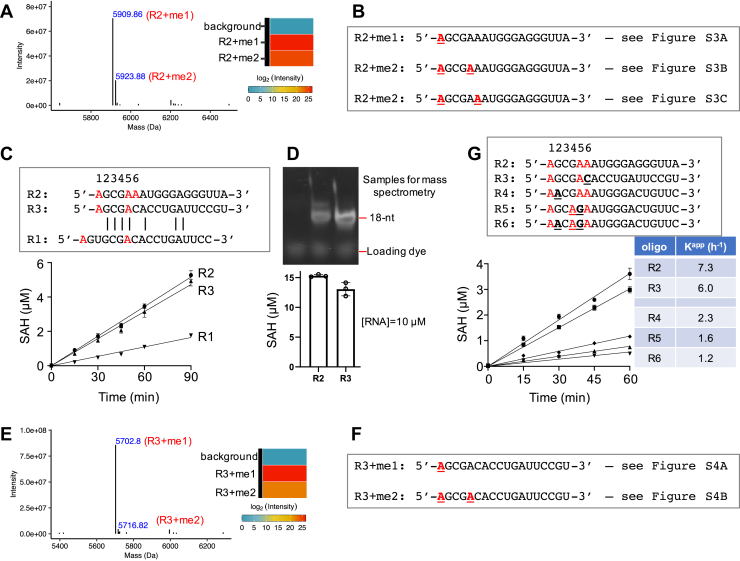
Figure 3.
PCIF1 activity on different RNA oligos.A, deconvoluted ESI spectra from intact mass spectrometry analysis of methylated oligo R2. Aggregated intensity corresponding to each methylation peak is shown to the right. B, summary of methylation sites (A in bold and underlined) with the corresponding tandem mass spectra shown in Fig. S3. C, oligos R2 and R3 have similar activity with the shared 5′-AGCGA sequence, whereas R1 has five residues (as opposed to three) between the first two adenosines. D, saturated reactions on oligos R2 and R3 (bottom) (N = 3). A 15% urea gel showing the methylated oligos used for mass spectrometry analysis (top). E and F, mass spectrometry analysis of methylated oligo R3. G, oligos R4, R5, and R6 have reduced activity, associated with substitutions at nucleotide positions 2, 4, and/or 5. ESI, electrospray ionization; PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1.