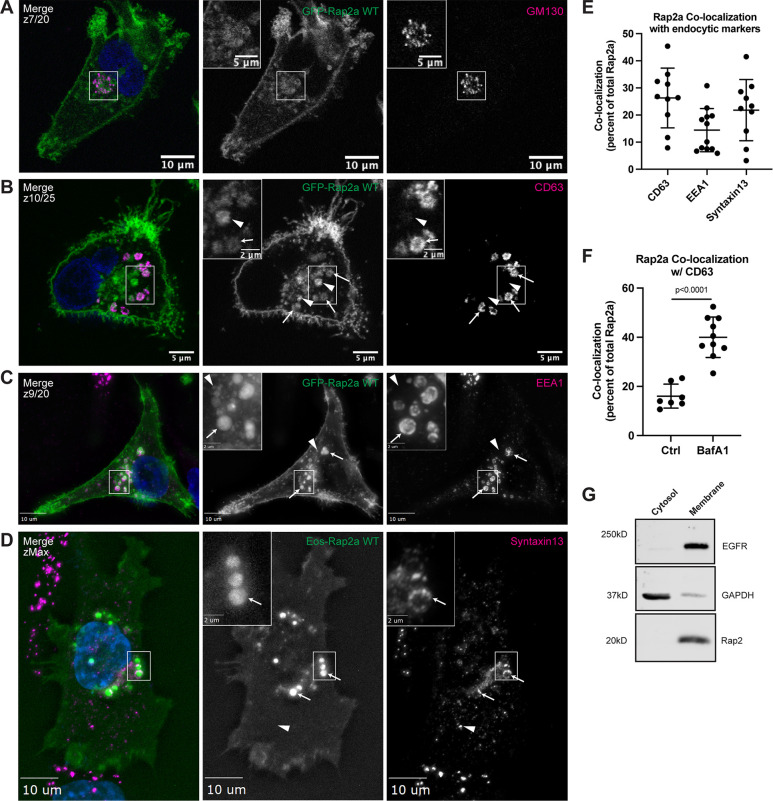
Figure 2.
Rap2 localizes to the plasma membrane and endolysosomal compartment in MDA-MB-231s. (A) Colocalization of GFP-Rap2a and GM130. MDA-MB-231 cells stably expressing GFP-Rap2a were fixed and stained with the Golgi marker GM130 (magenta) and DAPI (blue). Inset shows GFP-Rap2a and GM130 overlap. Scale bars, 10 and 5 µm. (B) Colocalization of GFP-Rap2a and CD63. MDA-MB-231 cells stably expressing GFP-Rap2a were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of GFP-Rap2a and CD63 overlap. Arrowheads point to GFP-Rap2a organelles that are not CD63 positive. Scale bars, 5 and 2 µm. (C) Colocalization of GFP-Rap2a and EEA1. MDA-MB-231 cells stably expressing GFP-Rap2a were fixed and stained with the early endosome marker EEA1 (magenta) and DAPI (blue). Arrows indicate examples of GFP-Rap2a and EEA1 overlap. Arrowheads point to GFP-Rap2a organelles that are not EEA1 positive. Widefield microscope. Scale bars, 10 and 2 µm. (D) Colocalization of Eos-Rap2a and Syntaxin13. MDA-MB-231 cells stably expressing Eos-Rap2a were fixed and stained with the recycling endosome marker Syntaxin13 (magenta) and DAPI (blue). Arrows indicate examples of Eos-Rap2a and Syntaxin13 overlap. Arrowhead points to Syntaxin13 endosome that is not Rap2a positive. Widefield microscope. Scale bars, 10 and 2 µm. (E) Rap2a colocalization analysis with endolysosomal compartments. 3i SlideBook6 software was used to calculate the percent of total GFP-Rap2a that colocalizes with the markers indicated (see Materials and methods). Two biological replicates were performed, with approximately five cells imaged for each replicate. Mean ± SD. n = 10 for GFP-Rap2a/CD63, n = 12 for GFP-Rap2a/EEA1, n = 10 for Eos-Rap2a/Syntaxin13. (F) Rap2a and CD63 colocalization analysis in Ctrl cells versus cells treated with lysosomal inhibitor Bafilomycin A1. MDA-MB-231 cells stably expressing GFP-Rap2a were treated with either DMSO (Ctrl) or 200 nM Bafilomycin A1 for 16 h. Cells were then fixed and stained for CD63, before colocalization analysis. 3i SlideBook6 software was used to calculate the percent of total GFP-Rap2a that colocalizes with CD63 (see Materials and methods). One biological replicate was performed. Mean ± SD. n = 7 cells for Ctrl, n = 10 cells for BafA1. Unpaired t test. P < 0.0001. (G) Cell fractionation analysis cytosol versus membrane. MDA-MB-231 parental cells were fractionated (see Materials and methods) to determine subcellular distribution of endogenous Rap2. Cytosol and membrane fractions were collected, separated by SDS-PAGE, and subjected to Western blot against known cytosol (GAPDH) and membrane (EGFR) markers. 30 µg of sample was loaded for each fraction. Source data are available for this figure: SourceData F2.