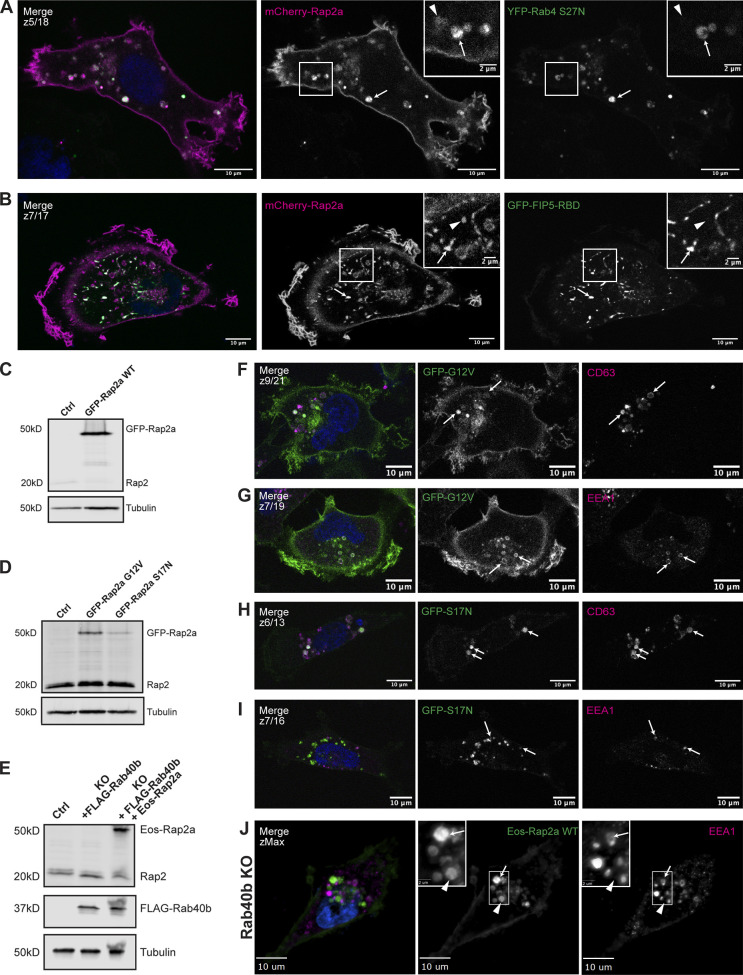
Figure S1.
Further characterization of Rap2a subcellular localization in MDA-MB-231 cells and analysis of Eos-Rap2a in Rab40b KO cells. (A) Colocalization of mCherry-Rap2a and YFP-Rab4 S27N. MDA-MB-231 cells were transiently cotransfected with mCherry-Rap2a and YFP-Rab4 S27N, followed by fixation and staining for DAPI (blue). Arrows indicate examples of mCherry-Rap2a and YFP-Rab4 S27N overlap. Arrowheads point to mCherry-Rap2a organelles that are not Rab4 S27N positive. Scale bars, 10 and 2 µm. (B) Colocalization of mCherry-Rap2a and GFP-FIP5-RBD. MDA-MB-231 cells were transiently cotransfected with mCherry-Rap2a and GFP-FIP5-RBD, followed by fixation and staining for DAPI (blue). Arrows indicate examples of mCherry-Rap2a and GFP-FIP5-RBD overlap. Arrowheads point to mCherry-Rap2a organelles that are not FIP5-RBD. Scale bars, 10 and 2 µm. (C) Western blot showing stable overexpression of GFP-Rap2a WT in MDA-MB-231s (lentivirus). Ctrl cells are MDA-MB-231 parentals. 50 µg of lysate was loaded for each sample. (D) Western blot showing stable overexpression of GFP-Rap2a-G12V and -S17N in MDA-MB-231s (lentivirus). Ctrl cells are MDA-MB-231 parentals. 50 µg of lysate was loaded for each sample. (E) Western blot showing generation of rescue line in Fig. 6 E. Rab40b KO cells were first stably transfected with FLAG-Rab40b WT (lentivirus, second column). Then, these cells were transfected with Eos-Rap2a and flow sorted (lentivirus, third column, flow sort instead of selection). Ctrl cells are dox-inducible Cas9 MDA-MB-231s that were used to generate Rab40b KO CRISPR line. 50 µg of lysate was loaded for each sample. (F) Colocalization of GFP-Rap2a-G12V and CD63. MDA-MB-231 cells stably expressing GFP-Rap2a-G12V were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-G12V and CD63 overlap. Scale bars, 10 µm. (G) Colocalization of GFP-Rap2a-G12V and EEA1. MDA-MB-231 cells stably expressing GFP-Rap2a-G12V were fixed and stained with the early endosome marker EEA1 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-G12V and EEA1 overlap. Scale bars, 10 µm. (H) Colocalization of GFP-Rap2a-S17N and CD63. MDA-MB-231 cells stably expressing GFP-Rap2a-S17N were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-S17N and CD63 overlap. Scale bars, 10 µm. (I) Colocalization of GFP-Rap2a-S17N and EEA1. MDA-MB-231 cells stably expressing GFP-Rap2a-S17N were fixed and stained with the early endosome marker EEA1 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-S17N and EEA1 overlap. Scale bars, 10 µm. (J) Eos-Rap2a colocalization with EEA1 in Rab40b KO cells. Rab40b KO MDA-MB-231 cells stably expressing Eos-Rap2a were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Widefield microscope. Arrows indicate examples of Eos-Rap2a and EEA1 overlap. Arrowheads point to Eos-Rap2a organelles that are not EEA1 positive. Scale bars, 10 µm, 2 µm. Source data are available for this figure: SourceData FS1.