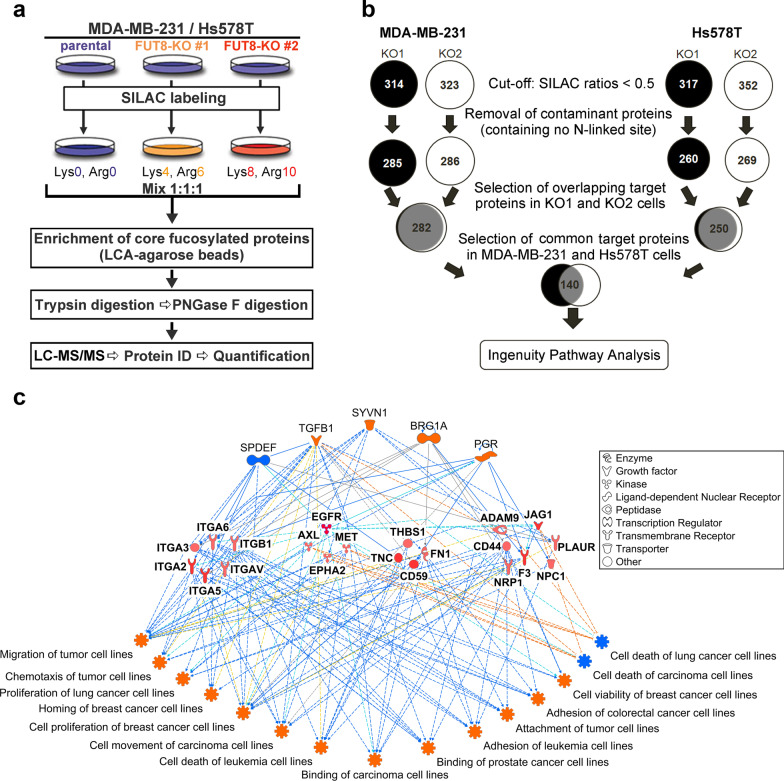
Fig. 2.
Quantitative glycoproteomics analysis. a Workflow of SILAC-based quantitative glycoproteomics in two highly invasive breast cancer cell lines (MDA-MB-231 and Hs578T). b Identification of core fucosylated glycoproteins. Using 0.5 as the threshold ratio, 282 and 250 candidate core-fucosylated glycoproteins were identified in MDA-MB-231 and Hs578T cells, respectively: 140 were common to both cell lines. c Functional network analysis of identified core fucosylated glycoproteins. Functional networks based on IPA predicted the activation state and subsequent effect on cellular functionality. Upstream regulators are in the top tier, and functions are in the bottom tier. FUT8 target proteins are in the middle tier. Orange lines indicate activation and blue lines suppression