Figure 4. Cryo-EM and nsEMPEM Fab-Env structures show apparent accommodation of gp120 N156 glycan after boosting.
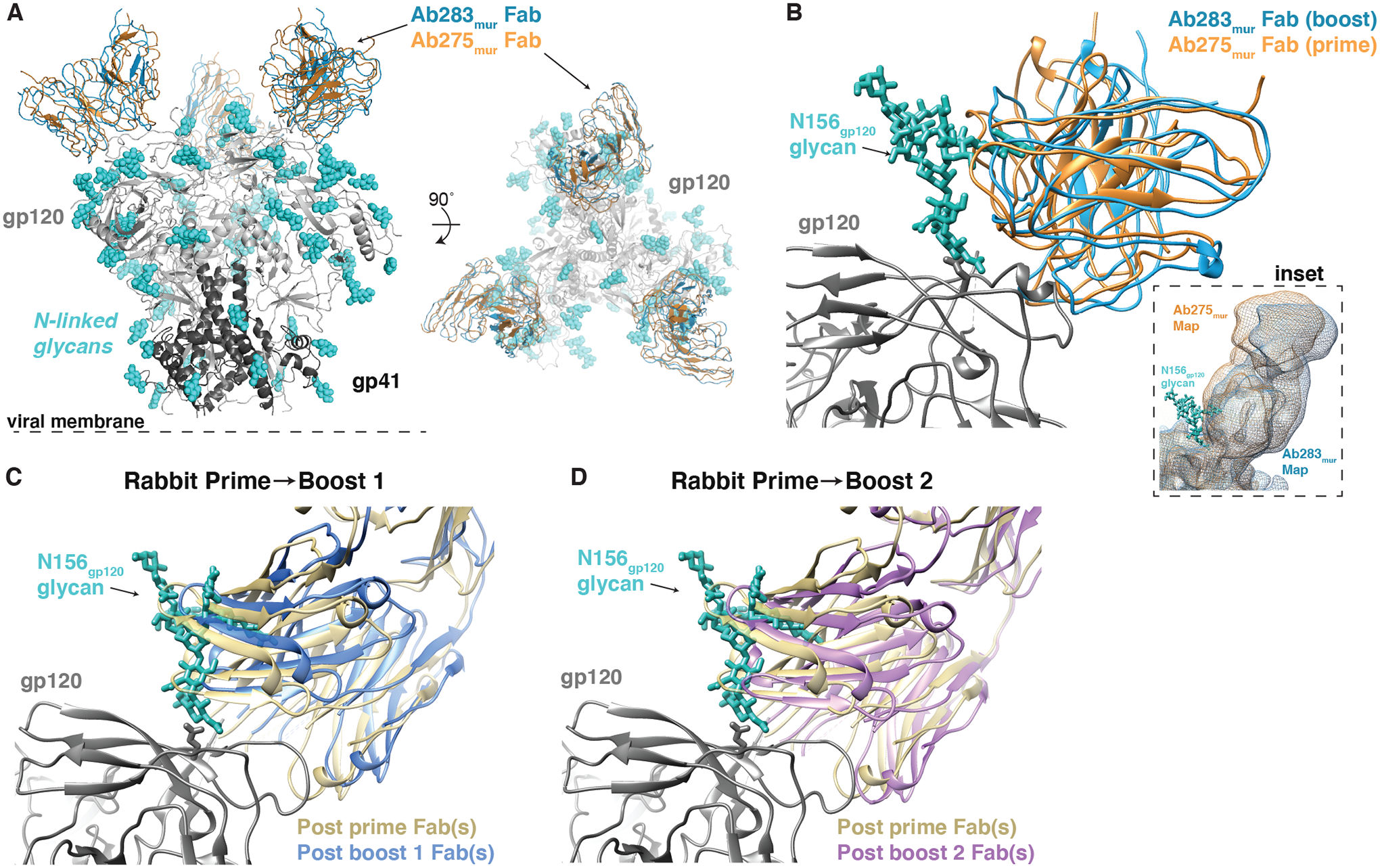
(A) A comparison of Ab275mur-RC1 (PDB 6ORQ) and Ab283mur-RC1 structures is shown. (B) A comparison of binding poses of two mouse monoclonal antibodies, Ab275mur and Ab283mur, isolated from wt mice after a prime only (Ab275mur) or prime plus sequential boost (Ab283mur) is shown. After aligning the Ab283mur-RC1 structure reported here and the Ab275mur-RC1 structure (PDB 6ORQ) on the RC1 coordinates, the pose of Ab283mur was shifted relative to that of Ab275mur in a direction that might better accommodate the gp120 N156 glycan (blue sticks from PDB 5T3X), which was present on boosting immunogens but not the priming immunogen. Inset: Overlay of low-pass filtered single-particle cryo-EM maps of the Fab-RC1 structures. (C and D) Coordinates docked into Fabs-Env nsEMPEM reconstructions (Fig. 2A) (C3 symmetry constraints applied) were aligned on the RC1 trimer and displayed as a close-up view of the antibody interaction site with the VH-VL domains of the post-P and post-B1 Fabs (C) or the post-P and post-B2 Fabs (D). The post-B1 Fabs and post-B2 Fabs appear to shift in a direction that might better accommodate the gp120 N156 glycan (blue sticks from PDB 5T3X).
