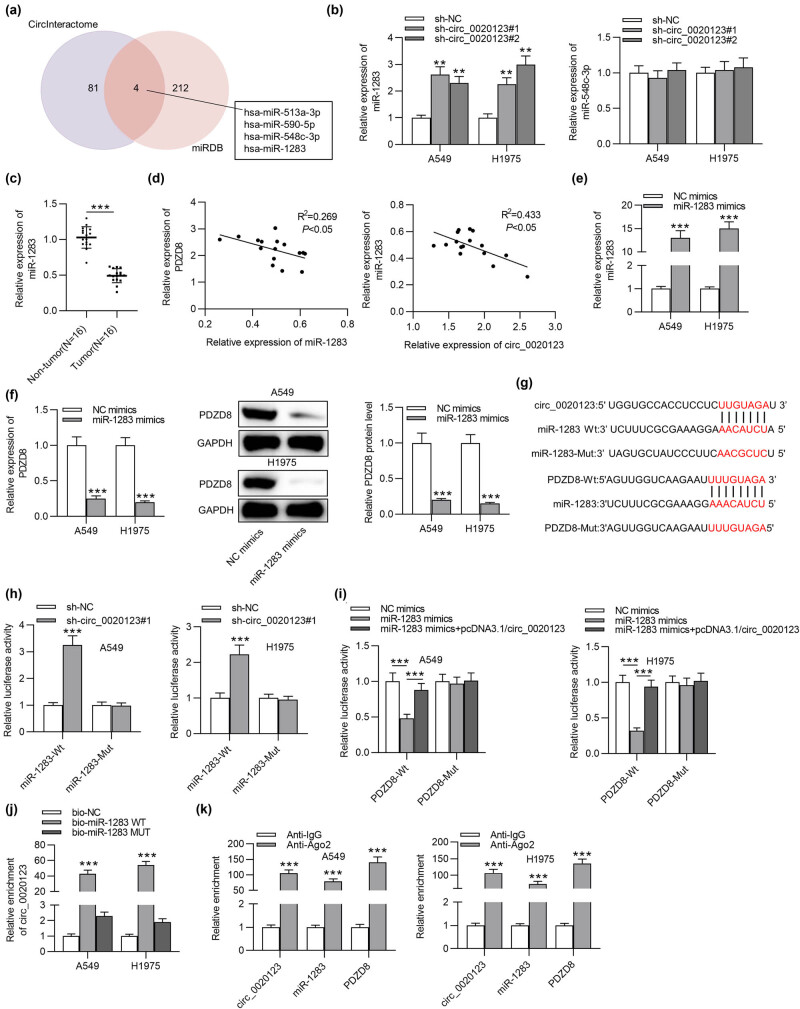
Figure 4.
circ_0020123 acts as a ceRNA against miR-1283 to regulate PDZD8. (a) CircInteractome and miRDB databases were utilized to predict miRNAs binding with circ_0020123 and PDZD8, respectively. (b) circ_0020123 knockdown upregulated miR-1283 expression and exert no significant effect on miR-548c-3p level in LUAD cells. **p < 0.01 vs sh-NC group. (c) MiR-1283 expression in LUAD tissues was relatively low. ***p < 0.001. (d) Based on Spearman correlation coefficient, miR-1283 expression was negatively correlated with circ_0020123 expression (R 2 = 0.269, *p < 0.05) and PDZD8 expression (R 2 = 0.433, *p < 0.05) in LUAD tissues. (e) miR-1283 expression was successfully overexpressed by miR-1283 mimics in LUAD cells. ***p < 0.001 vs NC mimics group. (f) miR-1283 overexpression decreased mRNA expression and protein levels of PDZD8 in A549 and H1975 cells. ***p < 0.001 vs NC mimics group. (g) The possible binding site between miR-1283 and circ_0020123 (or PDZD8) was predicted using CircInteractome and miRDB, respectively. (h) Luciferase reporter assays indicated the binding capacities between miR-1283 and circ_0020123. ***p < 0.001 vs sh-NC group. (i) The relationship of circ_0020123, miR-1283, and PDZD8 in LUAD cells has been verified by luciferase reporter assays. ***p < 0.001 vs NC mimics group or miR-1283 mimics group. (j) The direct interaction between circ_0020123 and miR-1283 was further confirmed by RNA pulldown assays. ***p < 0.001 vs bio-NC group. (k) The enrichment of circ_0020123, miR-1283, and PDZD8 in Ago2 and IgG antibodies was probed by RIP assays. ***p < 0.001 vs Anti-IgG group. The data are presented as the mean value ± standard deviation. Student’s t test was used to compare differences between LUAD tissues and normal tissues. One-way analysis of variance followed by Tukey’s post hoc analysis was used to compare differences among groups.