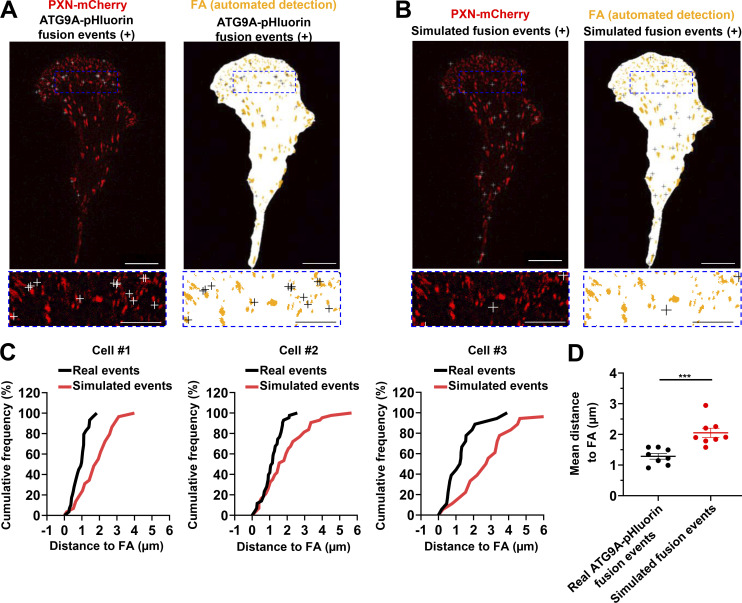
Figure 8.
ATG9A-positive vesicles target adhesion sites. U87 MG cells expressing PXN-mCherry and ATG9A-pHluorin were recorded using live-cell TIRF microscopy. (A) Left: Distribution of all observed ATG9A-pHluorin fusion events during the recording period (crosses), overlaid on the PXN-mCherry signal (red). Right: Derived synthetic image depicting the ventral cell surface area (white), the PXN-positive adhesion complexes (orange, automated detection using ImageJ), and the ATG9A-pHluorin fusion events (crosses). Scale bar, 20 µm; magnified views, 10 µm. (B) Left: Map of randomly simulated fusion events (crosses), overlaid on the PXN-mCherry signal (red). Right: Derived synthetic image depicting the ventral cell surface area (white), the PXN-positive adhesion complexes (orange), and the simulated fusion events (crosses). Scale bar, 20 µm; magnified views, 10 µm. (C) Cumulative frequency charts, from the cell shown in A (cell #1) and two other representative cells (cells #2 and #3), demonstrating the difference in distance to focal adhesions (FA) between real ATG9A-pHluorin fusion events (black line) and simulated events (red line). (D) Quantification of the mean distance to the centroid of closest focal adhesion for real ATG9A-pHluorin events or simulated events (n = 8 cells, for a total number of 365 events). Statistical significance was evaluated using Mann–Whitney U test. ***, P < 0.001.