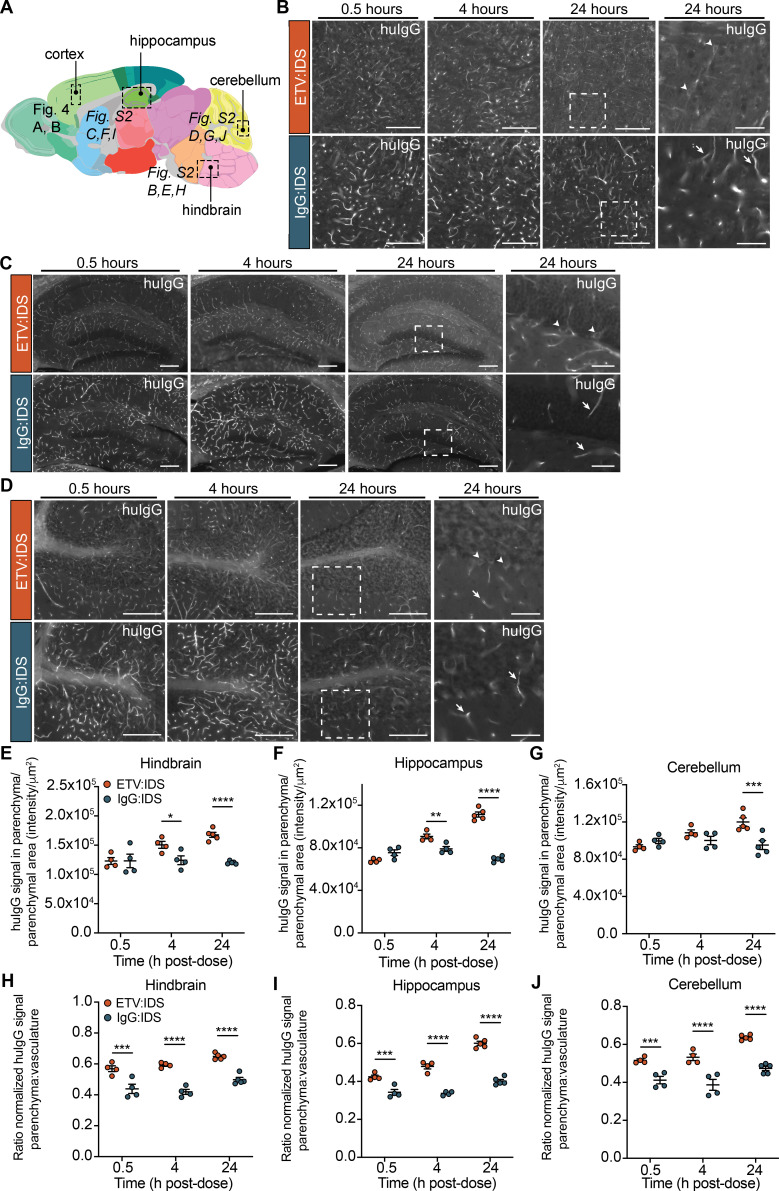
Figure S2.
IHC localization of ETV:IDS demonstrates more effective distribution into the brain parenchyma than IgG:IDS across multiple brain regions. The distribution of ETV:IDS and IgG:IDS was assessed using sagittal brain sections from TfRmu/hu KI mice 0.5, 4, and 24 h after an IV dose of 10 mg/kg. TfRmu/hu KI mice are on a C57BL/6 background. (A) Schematic indicating approximate location of sagittal brain regions of interest (ROIs) for images and quantification in Fig. 4, A and B (cortex), and Fig. S2 (hindbrain, hippocampus, and cerebellum; adapted from the Allen Adult Mouse Brain Atlas; original image credit: Allen Institute; Lein et al. [2007]). (B–D) Sections were immunostained with antibodies against huIgG, imaged, and post-stitched using a wide-field fluorescence slide scanner in the hindbrain (B), hippocampus (C), and cerebellum (D). Scale bars = 200 μm. Dashed boxes indicate regions shown at higher magnification displayed in the far-right panel; scale bars = 50 μm. Arrows indicate huIgG staining localized to vascular profiles; arrowheads indicate cellular internalization of huIgG staining. (E–G) Quantification of huIgG staining in the parenchyma of the hindbrain (E), hippocampus (F), and cerebellum (G) was calculated based on the total sum intensity of all parenchymal staining in the ROI divided by the total parenchymal area in the ROI. A custom macro script was used to identify blood vessels present in the tissue and masked out of subsequent image analyses. (H–J) The ratio of normalized huIgG signal in the brain parenchyma to brain vasculature was also quantified in the hindbrain (H), hippocampus (I), and cerebellum (J). Data presented are from a single study with n = 4–5 mice per group. Each symbol represents a biological replicate. Graphs display mean ± SEM and P values: two-way ANOVA with Sidak’s multiple comparison test; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.