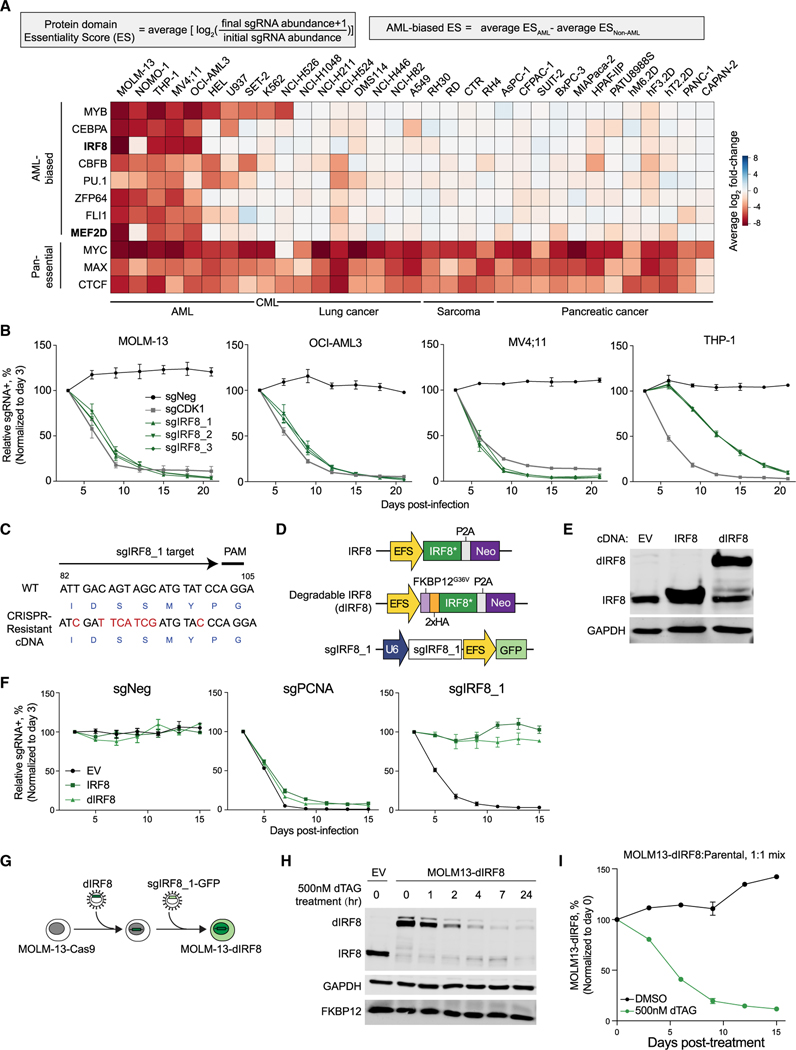
Figure 1. IRF8 is an AML-biased TF dependency.
(A) Summary of TF-domain-focused CRISPR screens. Genes were ranked by AML-biased ES defined by the difference in a particular domain’s ES in AML versus in non-AML cell lines. CML, chronic myeloid leukemia. Data are from Lu et al. (2018).
(B) Competition-based proliferation assays performed in indicated Cas9+ cell lines. sgRNA+ populations were monitored over time with a GFP co-expression marker. Plotted is the relative sgRNA+ population normalized to the day 3 sgRNA+ population over 21 days. sgNeg, negative control; sgCDK1, positive control.
(C) Design of CRISPR-resistant IRF8 cDNA. Encoded amino acids are labeled in blue at the bottom of the cDNA sequence.
(D) Vectors used for IRF8/dIRF8 cDNA complementation assay. IRF8*, sgIRF8_1-resistant IRF8 cDNA; dIRF8, degradable IRF8 that contains an additional FKBP12G36V domain and a 2×HA tag; Neo, neomycin resistance marker.
(E) Immunoblotting of FKBP12G36V-tagged IRF8, IRF8, or GAPDH (loading control) in whole-cell lysates of MOLM-13 cells transduced with indicated vectors.
(F) Competition-based proliferation assays performed in MOLM-13 Cas9+ cell lines stably expressing empty vector (EV), IRF8*, or dIRF8. sgPCNA, positive control.
(G) Schematic depicting establishment of an inducible IRF8 degradation system in MOLM-13 cells.
(H) Immunoblotting of whole-cell lysate of indicated cells treated with 500 nM dTAG-47 over time.
(I) Competition-based proliferation assay of MOLM-13-dIRF8 versus parental cells treated with either DMSO or 500 nM dTAG-47. Plotted is the relative sgRNA+ population normalized to the day 0 sgRNA+ population.
Data points of line graphs represent the average of three independent biological replicates (n = 3). Error bars represent mean ± SEM. See also Figure S1.