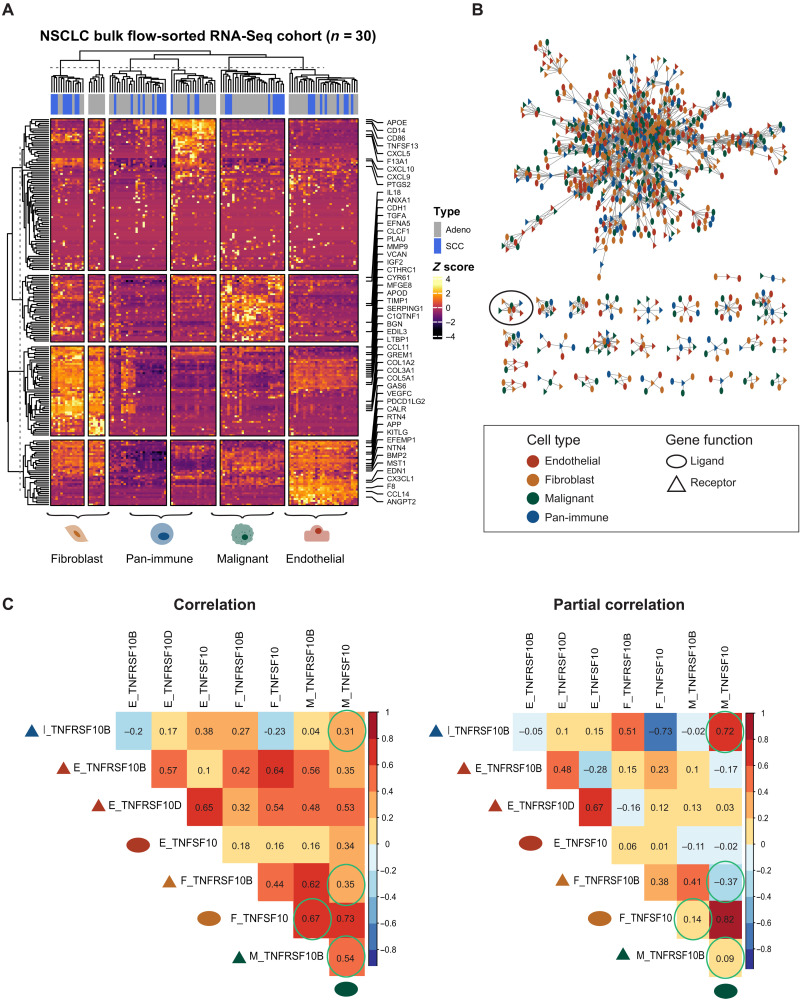
Fig. 1. Network representation of LTMI shows dense components of cognate LR pairs.
(A) All LR genes in the LUAD flow-sorted RNA-seq dataset hierarchically clustered. Genes labeled are the most highly expressed genes in the top five percentile. (B) Correlation network of expressed LR pairs from the Gentles et al. dataset. Color of nodes represent cell type of gene. Shape of node represents type of gene. Black circle indicates network component used for correlation analysis in panel below. (C) Correlation and partial correlation values of the nodes that are found within the black circled component in (B). Correlation matrix values are on the left, and partial correlation matrix values are on the right. The text labels of the correlation matrices correspond to the gene’s cell type added in front of the gene name (celltype_name). In addition, a colored (cell type) shape (gene function) is placed in front of the text label. Colors correspond to the legend in (B). Bottom dark green circle corresponds to M_TNFSF10 labeled at the top of the column. Green circles highlight examples of correlation values that are changed after accounting for confounding variables.