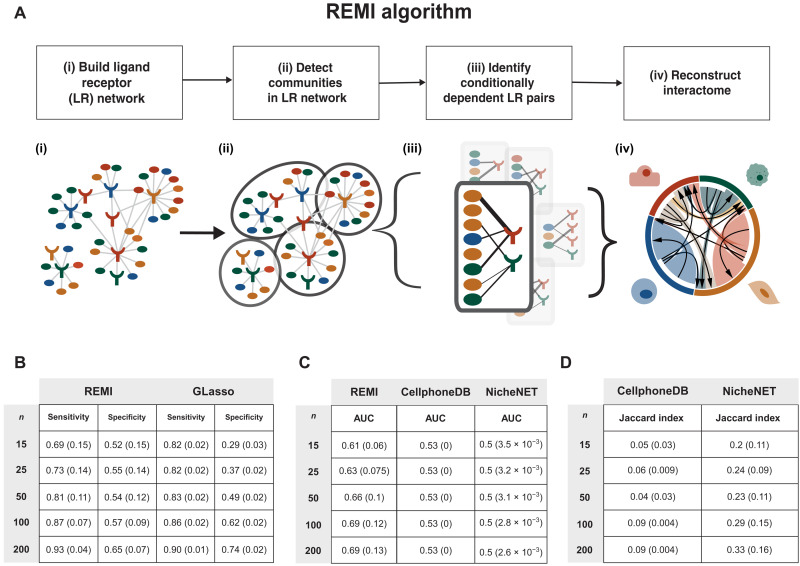
Fig. 2. Schematic overview of REMI and simulation studies.
(A) Diagram of REMI: (i) A network of potential LR interactions between cell types of interest is built given a reference LR database. Nodes represent either ligand or receptor genes. Edges represent known LR pairs. (ii) Network represented as density-based communities. (iii) Conditionally independent LR pairs removed: Each community is converted into a graphical Gaussian model, and conditionally independent LR pairs are removed. (iv) Conditionally dependent LR pairs across communities are reassembled into global interactome. (B) Simulation studies: Performance of REMI versus GLasso for sample sizes n = 15 to 200 when compared to a GLasso interactome derived from the entire LUAD TCGA RNA-seq dataset (n = 1031). Cohorts of various sample sizes were generated by downsampling TCGA data and then used to compare REMI’s and GLasso’s performance. At each sample size, 50 cohorts were generated. Mean AUC and SD (in parentheses) values are generated across the 50 cohorts per sample size. (C) Test performance of REMI compared with other computational cross-talk prediction algorithms. At each sample size, 50 cohorts were generated by randomly sampling TCGA data. Mean AUC and SD (in parentheses) are provided. (D) Average Jaccard indices of 50 cohorts per sample size calculated for each method compared to REMI.