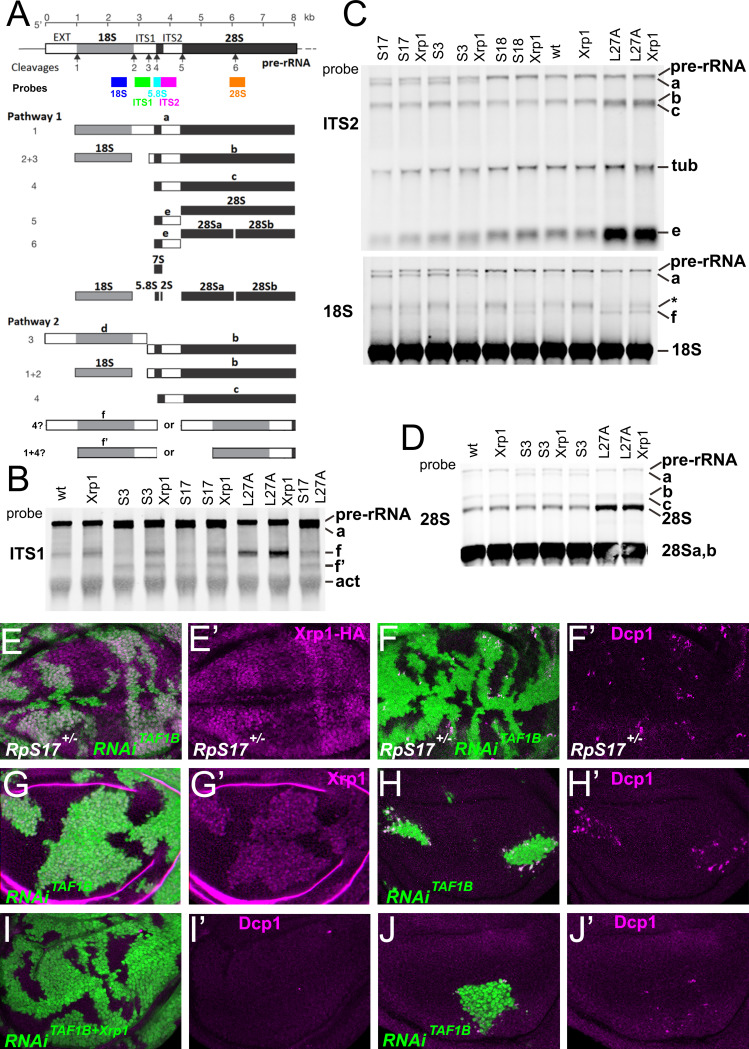
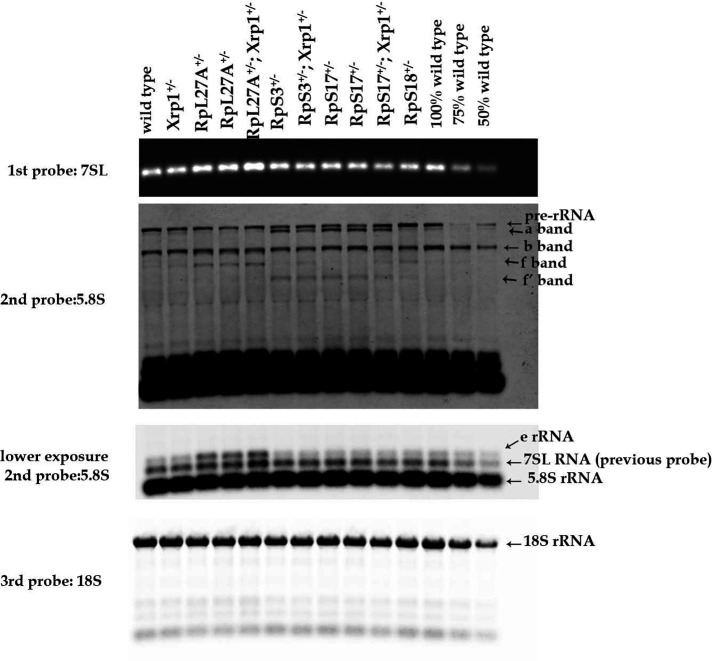
Figure 2. Ribosome biogenesis defects and their consequences.
(A) Two pathways of rRNA processing and the intermediates that result were characterized in D. melanogaster embryos by Long and Dawid. Mature 18 S, 5.8 S and 28Sa,b rRNAs are processed from the pre-RNA, along with the removal of two interval sequences ITS1 and ITS2. The cleavages sites were described by Long and Dawid. Colored boxes indicate the probes used in the present study. The 5.8 S probe overlaps with 147 bases at 3’of the ITS1 region, excluding cleavage site 3. Additional intermediates f and f’ were observed in the wing imaginal disc samples. These were recognized by ITS1, 5.8 S (Figure 2—figure supplement 1) and 18 S probe and therefore extended beyond the cleavage site 3, although whether beyond site four was uncertain. (B–D) Northern blots of total RNA purified from wild-type and Rp+/- wing discs, probed as indicated. (B) Reprobed with ITS1 after an initial actin probe. (C) Reprobed with ITS2 and then 18 S probes after an initial tubulin probe. Intermediates b, f and the 28 S rRNA (which in Drosophila is a precursor to the mature 28Sa and 28 Sb rRNAs) were detected in wild type and Xrp1+/- wing discs, other intermediates only in Rp+/- genotypes. RpS3+/- and RpS17+/- had lower levels of pre-RNA and intermediate (f) but accumulate intermediates (a) and (f’), which might indicate delays in cleavages 2 and 3. RpS18+/- had increased levels of pre-RNA and intermediate (f). RpL27+/- accumulated bands (b, c, e, and f) and 28 S. The effect on (f) suggests crosstalk between RpL27A and SSU processing. (E–I) show single confocal planes from mosaic third instar wing imaginal discs. (E) TAF1B depletion (green) increased Xrp1-HA levels in RpS17+/- discs (magenta, see also E’). (F) TAF1B depletion (green) increased in RpS17+/- discs led to cell death at the boundaries with undepleted cells (active Dcp1 staining in magenta, see also F’). (G) TAF1B depletion (green) also increased Xrp1 protein levels in RpS17+/+ discs (magenta, see also G’). (H) TAF1B depletion (green) led to cell death at the boundaries with undepleted cells (active Dcp1 staining in magenta, see also H’). (I) Co-depletion of Xrp1 with TAF1B (green) largely abolished cell death at the clone interfaces (active Dcp1 staining in magenta, see also I’). (J) Clones of cells depleted for TAF1B in parallel with panel I, showing reduced clones size and number (green), and competitive cells death at boundaries magenta, see also J’. Additional data related to this Figure is presented in Figure 2—figure supplement 1. Genotypes: Northerns: similar to Figure 1 and additionally: S17+/- genotype: p{hs:FLP}/ p{hs:FLP}; FRT42/+; FRT80 RpS17 p{arm:LacZ} /+, S17+/-; Xrp1+/- genotype: p{hs:FLP}/ p{hs:FLP}; FRT80 RpS17 p{arm:LacZ} /FRT82B Xrp1M2–73, S17+/-, L27A+/- genotype: p{hs:FLP}/ p{hs:FLP}; RpL27A- p{arm:LacZ} FRT40/+; FRT80 RpS17 p{arm:LacZ} /+, E, F: p{hs:FLP}/+; UAS- RNAiTAF1B /+;RpS17, act> CD2> Gal4, UAS-GFP /+ (line: v105873), G, H: p{hs:FLP}/+; UAS- RNAiTAF1B /+;act> CD2> Gal4, UAS- GFP /+ (line: Bl 61957), I: p{hs:FLP}/+; UAS- RNAiTAF1B /UAS-RNAiXrp1;act> CD2> Gal4, UAS- GFP /+ (line: Bl 61957), J: p{hs:FLP}/+; UAS- RNAiTAF1B /TRE-dsRed;act> CD2> Gal4, UAS- GFP /+ (line: Bl 61957) (processed in parallel with 2I).