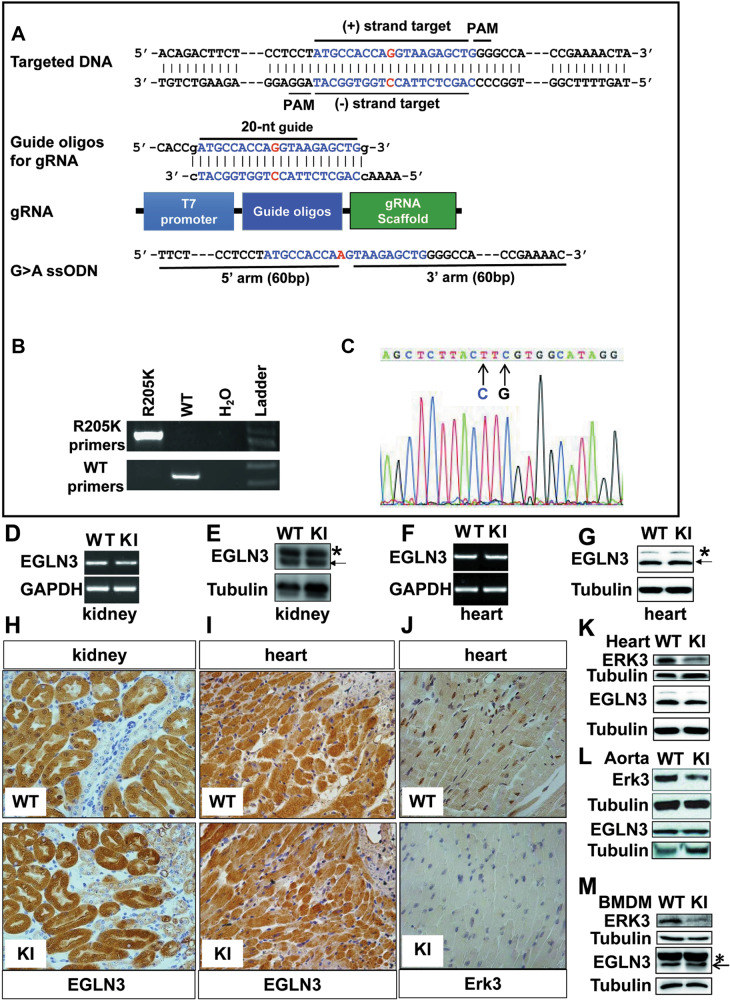
Fig. 1. Generation and identification of mouse strain deficient for EGLN3 hydroxylase activity.
A Shown is the strategy for the generation of EGLN3R205K knock-in (KI) mouse strain using CRISPR/Cas9 approach. B Genotyping of tail genomic DNA from wild-type or KI mouse by conventional PCR (n = 3). C DNA sequencing of genomic DNA from KI mouse (n = 2). D, F RT-PCR analysis of EGLN3 and R205K mRNA levels in the kidney (D) and heart (F) from wild-type or KI mouse (n = 3). E, G Immunoblotting analysis of EGLN3 and R205K protein levels in the kidney (E) and heart (G) from wild-type or KI mouse (n = 3). H, I Immunohistochemistry analysis of EGLN3 and R205K expression in the kidney (H) and heart (I) from wild-type or KI mouse (n = 3). J Immunohistochemistry analysis of Erk3 levels in the heart of wild-type or KI mouse (n = 3). K–M Immunoblotting analysis of Erk3 expression in the heart (K), aorta (L) and bone marrow-derived macrophages (BMDM) (M) of wild-type or KI mouse. GAPDH and Tubulin were used as the loading control (n = 3). CRISPR clustered regularly interspaced short palindromic repeats; ssODNs single-stranded DNA oligonucleotides; PAM protospacer adjacent motif; gRNA guide RNA; WT wild-type; KI knock-in; BMDM bone marrow-derived macrophages.