Figure 1.
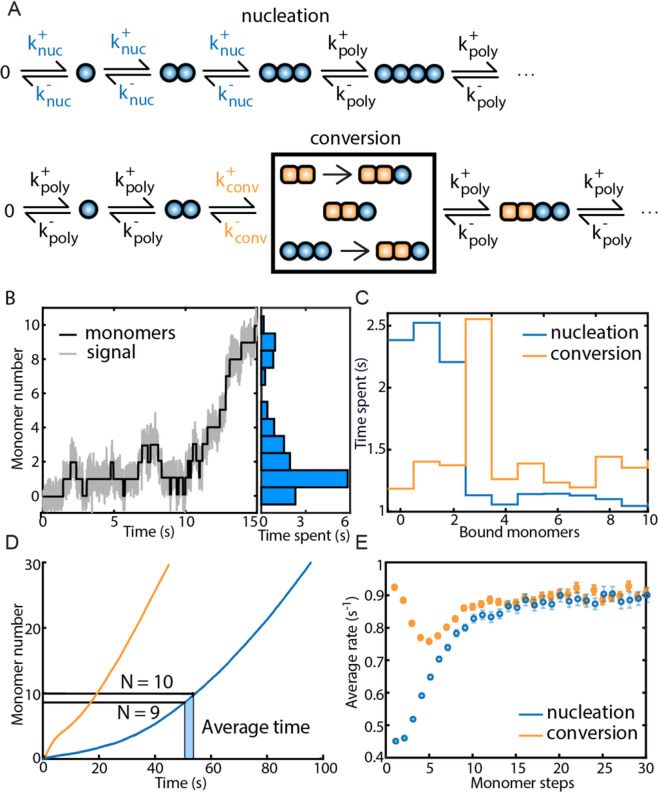
Analysis methods for single-filament data of protein self-assembly. (A) Kinetic model of the simulated nucleation and conversion process. For nucleation, k+nuc and k−nuc are changed to k+poly and k−poly as soon as the nucleus size is reached (here three monomers). For a conversion mechanism, only the assembly step associated with conversion (i.e., for example a restructuring of the assembled oligomer) shows k+conv and k−conv. Earlier and later steps follow k+poly and k−poly. For the conversion step, three main possibilities could lead to the slower kinetics as indicated by the black box: (1) the pre-existing oligomer needs to rearrange prior to the next binding event, (2) the restructuring occurs upon binding or (3), the restructuring occurs after binding of the next monomer. All possibilities result in a slower kinetic step and are treated the same in the simulations. (B) Example trace of a simulated growth process undergoing a nucleation mechanism with a nucleus size of four monomers (black) with Gaussian noise (grey). The distribution of time spent at a certain oligomer size (blue histogram) contains information about the nucleation mechanism when it is applied to many single oligomer traces (C). (C) Visitation analysis on a nucleation (blue) or a conversion mechanism (yellow). The visitation analysis identifies the nucleation mechanism as well as the nucleus size for nucleation or conversion. (D, E) Average rates analysis. (D) Average traces of a nucleation (blue) or conversion (yellow) mechanism. From the average trace of many individual filaments growing from the same starting point, average rates can be calculated from the average time it takes to reach the next oligomer size. (E) Average rates plotted versus the monomer number (oligomer size). Different nucleation mechanisms show individual signatures. The nucleation and conversion kinetics were simulated to show slower association rates than the polymerization kinetics.