Fig. 3.
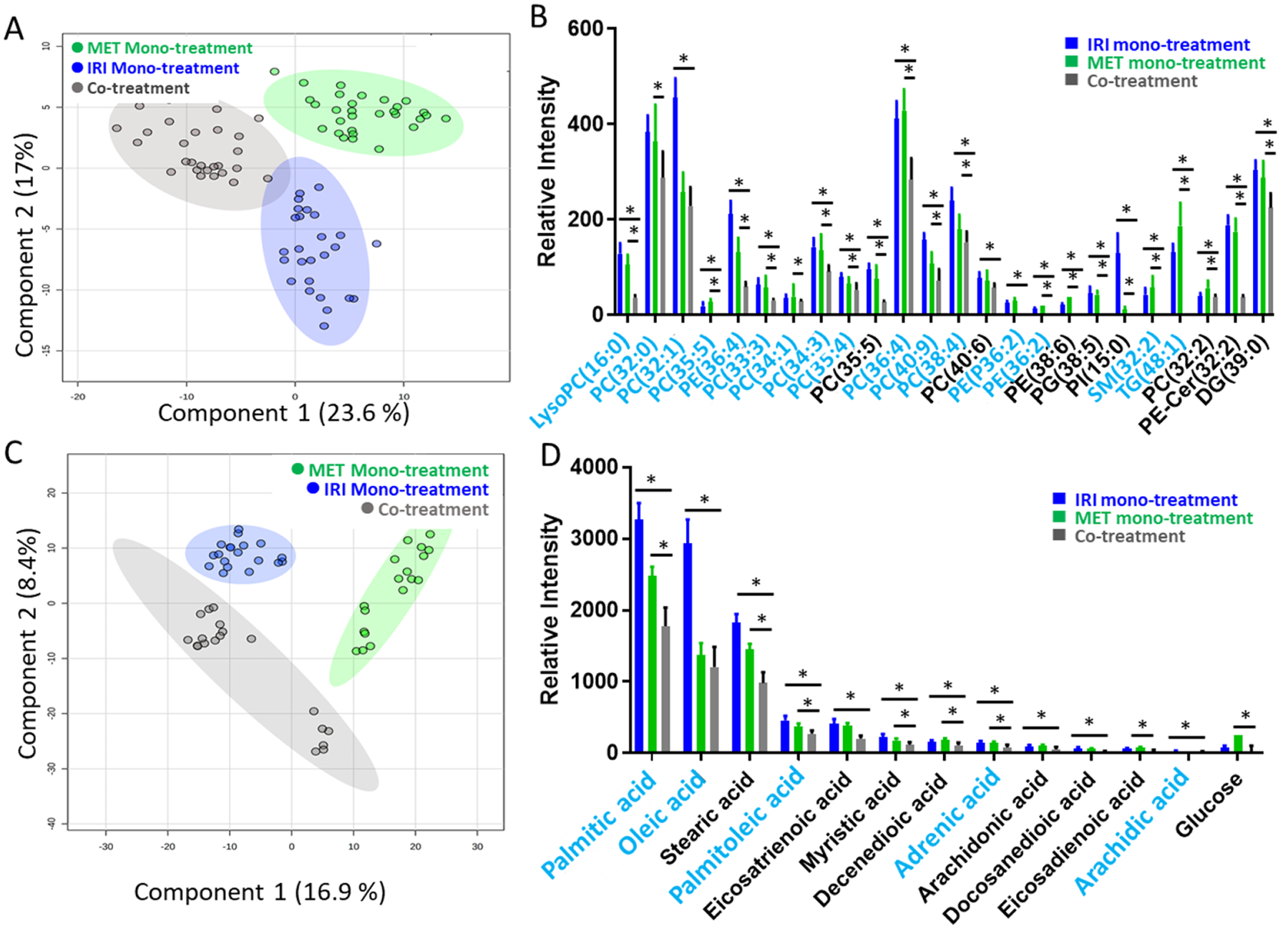
SCMS results of IRI-resistant cells from the co-treatment (0.56 μM IRI + 4.0 mM MET), IRI mono-treatment (22 μM IRI), and metformin mono-treatment (8.7 mM MET). (A) PLS-DA of positive ion mode results shows the metabolomics profiles were significantly changed by three different treatments (p < 0.001 from permutation test), and (B) representative lipids with significant abundance change (* p < 0.05 from both ANOVA and Tukey’s test) among three treatment groups. (C) PLS-DA of negative ion mode results shows metabolomics profiles were significantly changed by three different treatments (p = 0.004 from permutation test), and (D) representative fatty acids with significantly different abundances (* p < 0.05 from both ANOVA and Tukey’s test) among three treatment groups. Species labeled in blue font were identified using MS2 analyses using both single cells and cell lysates. (LysoPC: lysophosphatidylcholine; PC: phosphatidylcholine; PE: phosphatidylethanolamine; PI: phosphatidylinositol; PE-Cer: ceramide phosphoethanolamine; PG: phosphatidylglycerol; DG: diglyceride).
