Figure 8.
LVs encoding the U6-driven RNAi effectors produce a wide range of sequence-dependent and sequence-independent effects
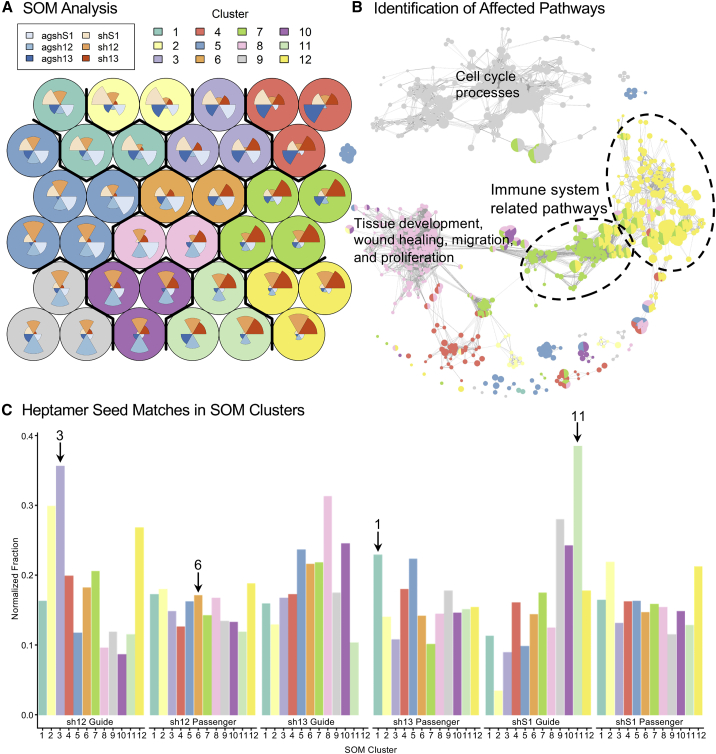
(A) Self-organizing map (SOM) analysis of the 3,000 most differentially expressed genes among differentially expressed genes in pairwise comparisons in the ARPE19 cells transduced with the LVs encoding the U6-driven RNAi effectors. Genes with similar expression profiles are grouped in the circular nodes. Nodes with similar profiles are organized in clusters. The polar area charts inside the nodes represent the expression profile of the genes in the node across the different samples. (B) Enrichment plot showing the enriched gene ontology biological processes (GO-BP) terms in the clusters from the SOM analysis. Captions for enriched pathways in selected clusters are shown (see Table S1). (C) Genes that contain heptamer seed site matches (HSMs, matching nt 2–8 of the RNAi effector strand) in their 3′UTR were identified for each predicted guide and passenger strand. The fraction of genes containing HSMs relative to both the number of genes in the SOM cluster and the total number of HSMs in the expressed genes was calculated for each SOM cluster and strand (normalized fraction). Arrows designate bars mentioned in the text.